Behavioural Genetic Interactive Modules
 Multivariate AnalysisOverviewThis module aims to demonstrate some of the principles underlying multivariate genetic analysis using twins. The ACE model, introduced in previous modules, is extended to the bivariate case: analysing two variables to decompose the covariance between measures into genetic and environmental components. TutorialThe Multivariate Analysis module
consists of three main panels. The panel at the top of the display
contains sliders which control nine parameters of the bivariate
ACE model. Adjusting these parameters will cause the Expected
correlations and Bivariate statistics (displayed in
the two panels below) to vary.
The aim of multivariate analysis would be to determine these nine parameters based on comparison between the three types of multivariate twin correlation:
This process is reversed in the current module: we can specify the nine parameters to determine what type of correlation structure we would expect to observe for different underlying aetiologies.
The elements of this correlation matrix are colour-coded (as are the other two correlation matrices). The top diagonal element is coded red as it represents Trait 1. The second diagonal element is coded green as it represents Trait 2. The off-diagonal element is coded white as it represents a correlation between the two traits. An important point to note, however, is that the "white" elements of all three expected correlation matrices are determined not just by the three Correlation parameters in the panel above: they will also depend on the values of the six univariate parameters. Exploring the modules, you will be able to examine these relationships further.
ExampleUse the module to verify the example of multivariate analysis in the Appendix, where three correlation matrices were given (phenotypic, MZ and DZ) from which the univariate statistics, the genetic and environmental correlations and the bivariate statistics were calculated. Although the example was for three traits the module can only deal with two at a time, the bivariate case. (Multivariate analysis can, in principle, deal with any number of variables.) However, it is perfectly valid to extract the appropriate pairwise values for only two of the three traits and we should expect to find a similar pattern. Instead of calculating the parameters given the observed correlations, we can use the module to calculated the expected correlations, given certain parameter values: this should give similar results. Parameters Univariate statistics
Expected Phenotypic correlations
For example, to confirm the relationship between X and Y, enter the relevant univariate parameters as Trait 1 and Trait 2, enter the genetic, shared environmental and nonshared correlations between X and Y and confirm that the expected correlations are similar to those observed in the example. Note the bivariate statistics. What conclusions can you draw from these about the relationships between these three traits?
QuestionsQuestion 1. Using the module, do you notice any special relationship between the three bivariate statistics and the phenotypic correlation between Trait 1 and Trait 2? If so, why does this relationship exist?
Question 2. Is there any scenario in which we might observe little or no correlation between the two traits despite a high genetic correlation? Verify your answer using the module.
Question 3. Is there any scenario where we might expect DZ twins to be more similar than MZ twins? Verify your answer using the module.
Answers
Answer 1. The bivariate statistics always sum to the phenotypic correlation. This is because they represent the proportion of between-trait shared variance which is shared due to either additive genetic, shared environmental or nonshared environmental influences. The phenotypic correlation, by definition, represents the sum of these three sources of covariation between the two traits.
Answer 2. If two traits have a high positive genetic correlation but a negative (non)shared environmental correlation (or vice versa) then we might expect to find near zero phenotypic correlations (depending on the relative balance of genetic and environmental influences for the two traits). This point is important because it shows that two traits that appear phenotypically unrelated might actually share genetic or environmental causes.
Answer 3. We might expect DZ twins to show a higher cross-trait, cross-twin correlation than MZ twins if both traits are strongly heritable and there is a negative genetic correlation. This is because a negative genetic correlation implies that genetic influences that increase one trait will tend to decrease the other trait. Under this scenario, we might find that MZ twins show a cross-trait, cross-twin correlation of, say -0.2, whilst DZ twins show a correlation of -0.1. In one sense, DZ twins have the higher correlation. However, if a correlation is negative, then a lower value (i.e. closer to -1) implies a stronger relationship between the two traits. In this sense, MZ twins are still more similar to each other than DZ twins (despite the correlations being numerical lower). There are scenarios where absolute value of the DZ cross-trait correlation can be greater than MZ cross-trait correlation. Try setting the both traits to half additive genetic, half shared environmental influence and set the genetic correlation to -0.8 and the shared environmental correlation to 0.9. Whether such situations are ever likely to occur in practice is another matter, but certainly the model allows for such possibilities. (Note that the cross-twin, same-trait correlations can never be greater in DZ twins than in MZ twins under the basic ACE model.)
Return
to Modules Menu
|
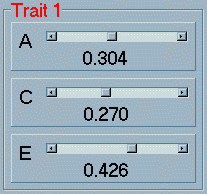 The nine parameters of the bivariate ACE
model are divided into three colour-coded groups: three univariate
parameters for the first trait (in red) which represent the additive
genetic, shared environmental and nonshared environmental components
of variance. Note that these parameters are expressed as proportions
of variance - that is, they will always sum to 1 (which is
why increasing one slider will cause the two other values to
drop slightly).
The nine parameters of the bivariate ACE
model are divided into three colour-coded groups: three univariate
parameters for the first trait (in red) which represent the additive
genetic, shared environmental and nonshared environmental components
of variance. Note that these parameters are expressed as proportions
of variance - that is, they will always sum to 1 (which is
why increasing one slider will cause the two other values to
drop slightly).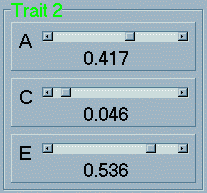 In
a similar manner, there are three univariate parameters that
can be specified for the second trait (coded in green). In this
case, the heritability of Trait 2 is set to just under
42%, the shared environment to just under 5% and the nonshared
environmental to just under 54%. Note: you may find it
difficult when trying to specify exact values for the three parameters
because of the inter-dependency amongst them that makes them
all change when only one is changed (to ensure they always sum
to 1, as mentioned).
In
a similar manner, there are three univariate parameters that
can be specified for the second trait (coded in green). In this
case, the heritability of Trait 2 is set to just under
42%, the shared environment to just under 5% and the nonshared
environmental to just under 54%. Note: you may find it
difficult when trying to specify exact values for the three parameters
because of the inter-dependency amongst them that makes them
all change when only one is changed (to ensure they always sum
to 1, as mentioned).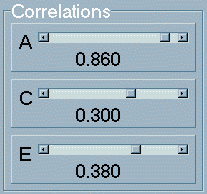 The
remaining three parameters represent the correlations
between univariate parameters (coded white). The concept of a
genetic, shared environmental or nonshared environmental correlation
is explained in the Appendix. As these parameters are
correlations, not proportions of variance, they range from -1
to +1 instead of 0 to 1. A high, positive genetic correlation
would imply that the genetic influences on one trait tend to
overlap with the genetic influences on the second trait, independent
of the actual magnitude of the genetic influence for either trait.
The
remaining three parameters represent the correlations
between univariate parameters (coded white). The concept of a
genetic, shared environmental or nonshared environmental correlation
is explained in the Appendix. As these parameters are
correlations, not proportions of variance, they range from -1
to +1 instead of 0 to 1. A high, positive genetic correlation
would imply that the genetic influences on one trait tend to
overlap with the genetic influences on the second trait, independent
of the actual magnitude of the genetic influence for either trait.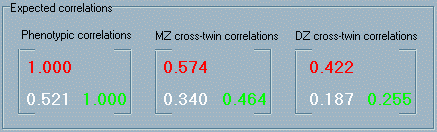 The
three types of correlation above are expressed, in a slightly
different manner, in the Expected correlations panel of
the module. This panel represents three correlation matrices.
The first matrix is labelled Phenotypic correlations and
represents the within-individual correlations. Of course, within-individual,
within-trait correlations will always be 1 - these are represented
by the diagonal elements of this matrix. The off-diagonal element
is of primary interest in this matrix: it represents the cross-trait,
within-individual correlation. That is, this is the correlation
we would expect to find between Trait 1 and Trait 2
if we measured both traits in a large number of unrelated individuals
to see how the one individual's score on Trait 1 is associated
with their score of Trait 2.
The
three types of correlation above are expressed, in a slightly
different manner, in the Expected correlations panel of
the module. This panel represents three correlation matrices.
The first matrix is labelled Phenotypic correlations and
represents the within-individual correlations. Of course, within-individual,
within-trait correlations will always be 1 - these are represented
by the diagonal elements of this matrix. The off-diagonal element
is of primary interest in this matrix: it represents the cross-trait,
within-individual correlation. That is, this is the correlation
we would expect to find between Trait 1 and Trait 2
if we measured both traits in a large number of unrelated individuals
to see how the one individual's score on Trait 1 is associated
with their score of Trait 2.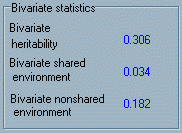 The
final panel represents the bivariate statistics. These
three statistics would often be calculated after multivariate
genetic analysis has been performed as they provide useful information
regarding the extent to which the phenotypic correlation between
two traits is mediated by genetic and/or environmental factors.
For example, the bivariate heritability is a measure of the extent
to which shared genetic influence generates a correlation between
two traits. If two traits had a phenotypic correlation of 0.5
and bivariate heritability of 0.4, we could conclude that 80%
of the correlation is mediated by shared genetic influence. Bivariate
heritability is therefore distinct from the genetic correlation.
For example, even if two traits have a very high genetic correlation,
if neither trait is strongly heritable then shared genetic influences
are unlikely to explain much of the observed correlation between
the two traits. The calculation of the bivariate heritability
is outlined in the Appendix : it is essentially a function
of the two univariate heritabilities and the genetic correlation.
The bivariate statistics for the shared and nonshared environment
are equivalent to the bivariate heritability in calculation and
interpretation.
The
final panel represents the bivariate statistics. These
three statistics would often be calculated after multivariate
genetic analysis has been performed as they provide useful information
regarding the extent to which the phenotypic correlation between
two traits is mediated by genetic and/or environmental factors.
For example, the bivariate heritability is a measure of the extent
to which shared genetic influence generates a correlation between
two traits. If two traits had a phenotypic correlation of 0.5
and bivariate heritability of 0.4, we could conclude that 80%
of the correlation is mediated by shared genetic influence. Bivariate
heritability is therefore distinct from the genetic correlation.
For example, even if two traits have a very high genetic correlation,
if neither trait is strongly heritable then shared genetic influences
are unlikely to explain much of the observed correlation between
the two traits. The calculation of the bivariate heritability
is outlined in the Appendix : it is essentially a function
of the two univariate heritabilities and the genetic correlation.
The bivariate statistics for the shared and nonshared environment
are equivalent to the bivariate heritability in calculation and
interpretation.