Luna: software for the analysis of sleep signal data
Luna is an open-source C/C++ software package for manipulating and
analyzing polysomnographic recordings, with a focus on the sleep EEG.
Originally oriented around command-line scripting
(lunaC), we are developing various extensions,
including the Python module
lunapi and the lunaR library
for the R statistical package. The
current release is v1.2.2 (7-Mar-2025): see here for
a list of changes/additions. Please direct any questions to luna.remnrem@gmail.com.
Luna in your browser
Test drive lunapi (the Python instantiation of Luna) in your
browswer, linked to example data and interactive notebooks using
this link to a Binder-hosted cloud
instance.
For more information on lunapi, see these
pages.
A family of Luna tools
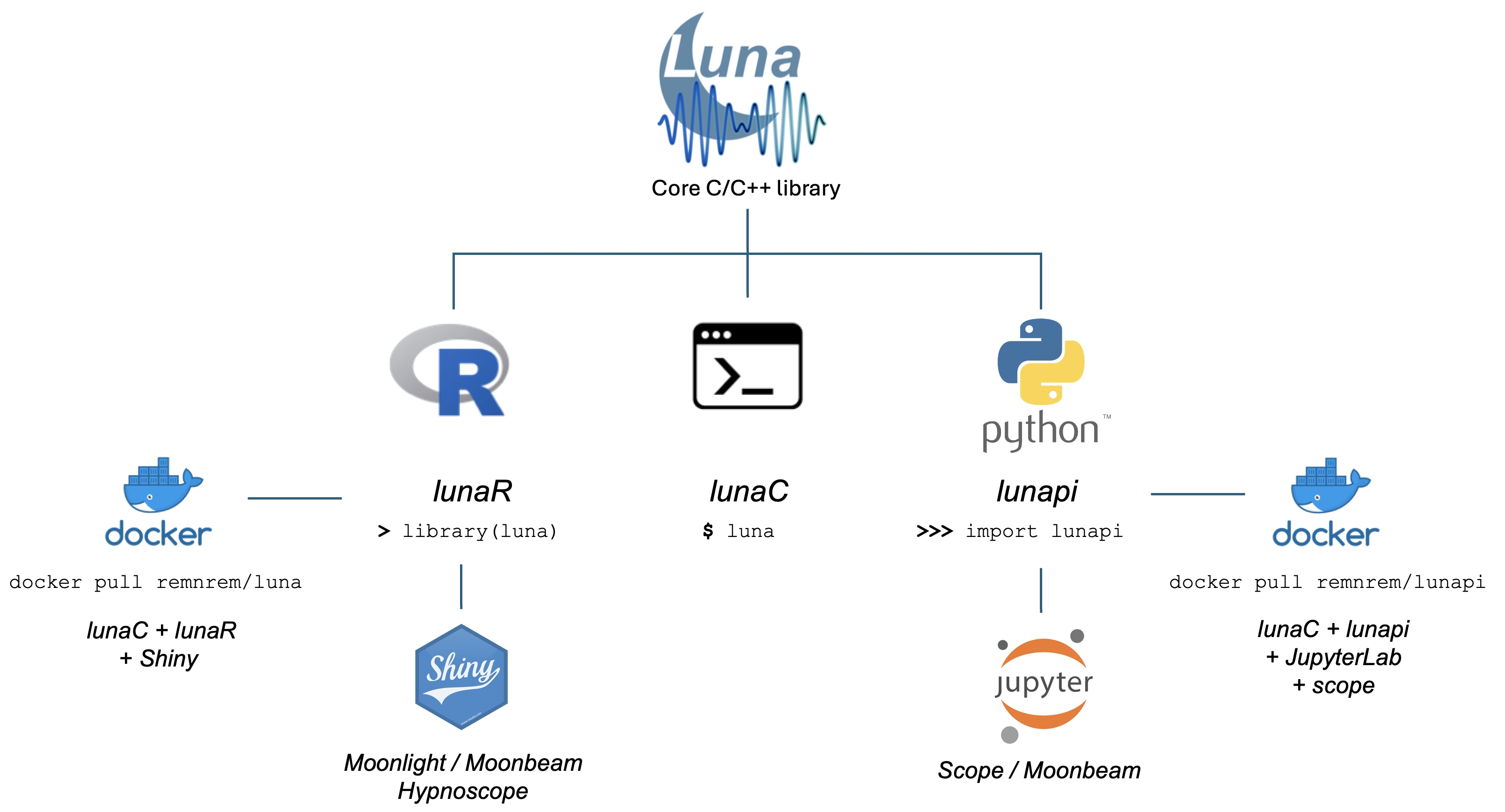
The Luna package comprises a number of related components, all of which are oriented around the same core C/C++ library. The core library implements all the commands for working with sleep signal data. There are three main ways to use the library, all of which provide fundamentally the same basic functionality:
-
via the lunaC command-line tool: this is the original interface, and is still the best approach for working with large datasets, ideally with scripted analyses and using a Unix-like cluster-computing environment
-
via the lunapi Python package: this is the best approach for interactive analyses and for those familiar with Python; you can try it out in your browser via a Binder-hosted notebook here
-
via the lunaR R library: users more familiar with R may prefer this option (although note that although we plan to maintain this as is, in the future more effort will be put into developing the Python interface)
Note
In this documentation, we often refer to lunaC simply as
Luna. Also, luna is the filename of both the actual command line
executable and the R package, and so we use the terms lunaC to disambiguate it from
lunapi or lunaR where necessary. Most material is
common to all packages, as they are based on the same basic C/C++ Luna
library.
On top of these components, we have also developed a few interactive tools:
-
Moonlight, an interactive viewer and web-based frontend, based on lunaR (and also available via http://remnrem.net
-
scope, an interactive viewer designed for the JupyterLab environment using lunapi
-
Moonbeam, a connector to NSRR data, available in either lunapi or lunaR
These tools can either be installed individually or they can be accessed via prebuilt Docker containers
Getting started
1. After downloading one or more of the Luna interfaces, the best place to start is the tutorial. The tutorial is initially couched in terms of the command-line tool, but versions are available for both R and Python interfaces.
2. You might then want to step through the walk-through, referencing this main documentation throughout (although note that completing the multi-part walk-through may take several sessions).
3. Then work your way through the pages listed in the left-hand side menu. (On devices with smaller screens this may be minimized: if so, click the top left three horizontal bars icon.) In particular, the Concepts page describes many key ideas and conventions, that are relevant for all implementations of Luna. The Commands page tabulates all primary Luna commands (these are common across command-line, R and Python implementations).
Things Luna aims to do
The Commands pages list all supported functionality; main areas are summarized below.
Primary use cases
- Read, manipulate and write large sets of EDF and EDF+ signals
- Filter, resample and re-reference signals
- Generate a variety of (per-epoch) summary statistics
- Statistical artifact detection for EEG channels
- Annotate and mask/filter epochs
- Estimate key features of sleep macro-architecture
- Automated sleep staging
- Spectral analyses
- Spindle and slow oscillation detection
- Coherence and cross-frequency coupling
- Multi-channnel, topographical analyses
- Interval-based analyses of event timing
- Sample-level linear association models
- Manipulating annotation data and meta-data
- Visual data exploration via Moonlight or scope
Things Luna doesn't aim to do
Luna was originally designed to work with the large number of polysomnograms at the NSRR, with a focus on intersecting sleep EEG signals with other annotations. As such, some areas are not well supported, or effectively outside of Luna's scope.
Areas outside of Luna's primary focus
-
Methods development platform: although the Python/R extensions can support methods development, other tools (including general purpose Matlab packages such as EEGLAB) will be better suited for expert users interested in flexibly altering and developing new analyses
-
Analyses of cardiac and respiratory events: most of Luna's specialized sleep analyses are currently focused on EEG signals (e.g. spindles and slow oscillations)
-
Online signal processing: Luna is set up for all analyses being done offline, i.e. on the entire recording
-
Support for multiple formats: currently, Luna is mainly based around EDF and EDF+ files (as well as plain text)