FIR filters
Linear-phase signal filters
| Command | Description |
|---|---|
FILTER |
Apply a FIR filter to one or more signals |
FILTER-DESIGN |
Display filter properties |
FILTER
Applies a linear-phase FIR filter to a signal
This command modifies the in-memory signal, by applying a finite impulse response (FIR) filter, which can be either a low-pass, high-pass, band-pass or band-stop filter.
Parameters
Core parameters are as follows:
| Parameter | Example | Description |
|---|---|---|
sig |
sig=C3,F3 |
Restrict analysis to these channels |
bandpass |
bandpass=0.3,35 |
Band-pass filter between 0.3 and 35 Hz |
lowpass |
lowpass=35 |
Low-pass filter with cutoff of 35 Hz |
highpass |
highpass=0.3 |
High-pass filter between 0.3 and 35 Hz |
bandstop |
bandstop=55,65 |
Band-stop filter between 0.3 and 35 Hz |
fft |
Use FFT to implement the filter (this is now the default) |
By default, the FILTER command uses the Kaiser window approach to
define the filter, which requires the two following parameters:
| Parameter | Example | Description |
|---|---|---|
ripple |
ripple=0.01 |
Ripple (as a proportion) |
tw |
tw=1 |
Transition width (in Hz) |
tw |
tw=0.5,5 |
Separate lower and upper transition widths for a bandpass filter (in Hz) |
Note that if using bandpass tw=lwr,upr then you need to specify two values for ripple also (althogh
these can be similar).
Ripple and stopband attenuation
The ripple parameter is related to stopband attenuation by the relation
A = -20log10(ripple), such that a ripple=0.01 corresponds to -40 dB stopband attenuation.
Alternative filters
Alternatively, it is possible to
-
read the FIR coefficients in from a
file -
use the window method to design a FIR with a fixed filter order, using either a Bartlett, Hann, Blackman or rectangular window
-
use a narrow-band Gaussian filter, defined by a center frequecy and FWHM (full-width half-maximum) bandwidth, with
ngaus -
use an IIR Buttworth or Chebyshev filter
| Parameter | Example | Description |
|---|---|---|
file |
file=fir1.txt |
Read FIR coefficients from a file |
order |
order=30 |
Fix the FIR filter order |
rectangular |
Specify a rectangular window | |
bartlett |
Specify a Bartlett window | |
hann |
Specify a Hann window | |
blackman |
Specify a Blackman window | |
ngaus |
ngaus=12,2 |
Specify a narrow-band Gaussian filter (center frequency, FWHM) |
butterworth |
butterworth=4 |
A Butterworth IIR filter of fixed order (see below) |
chebyshev |
chebyshev=4,1 |
A Chebyshev IIR filter of fixed order and ripple factor (see below) |
IIR filters
Butterworth and
Chebyshev (type I)
filters require one of bandpass=f1,f2, bandstop=f1,f2,
lowpass=f1 and highpass=f1 options. Butterworth takes a single
parameter, the filter order; Chebyshev takes two: order and the ripple
factor epsilon - see the above link for a definition of how the ripple
factor relates to passband ripple in dB. Chebyshev filters are
sharper than the Butterworth filter, whereas Butterworth filters
control passband ripple. Note that both IIR introduce phase delays:
in general, FIR filters are probably preferable choices.
Output
After running FILTER, the in-memory signal for a filtered channel
will represent the filtered signal. No explicit output is generated
by the FILTER command.
Examples
As a first example, here we apply a bandpass filter to the EEG channel
C3, with transition frequencies of 0.3 and 35 Hz, using a FIR filter
designed via the Kaiser window method with a ripple parameter
(ripple) of 0.01 and a transition width parameter (tw) of 0.5 Hz:
FILTER sig=C3 bandpass=0.3,35 ripple=0.01 tw=0.5
In this second example, we use lunaR to copy a
signal, apply different bandpass filters to each duplicate
(corresponding to delta, theta, alpha, sigma and beta frequency bands)
and then plot the results. Here we use the individual nsrr02 from
the tutorial dataset. In R, where we assume that the
current directory is the one in which we unzipped the tutorial dataset
(i.e. and so the s.lst sample-list is present), we'd enter the following:
library(luna)
lattach( lsl( "s.lst" ) , "nsrr02" )
nsrr02 : 14 signals, 10 annotations, 09:57:30 duration
EEG using SIGNALS:
leval( "SIGNALS keep=EEG" )
nsrr02 : 1 signals, 10 annotations, 09:57:30 duration
We then duplicate this signal using COPY
five times, giving new labels to each:
leval( "COPY sig=EEG tag=DELTA" )
leval( "COPY sig=EEG tag=THETA" )
leval( "COPY sig=EEG tag=ALPHA" )
leval( "COPY sig=EEG tag=SIGMA" )
leval( "COPY sig=EEG tag=BETA" )
nsrr02 : 6 signals, 10 annotations, 09:57:30 duration
lchs(), we confirm that all channels have been appropriately added:
lchs()
[1] "EEG" "EEG_DELTA" "EEG_THETA" "EEG_ALPHA" "EEG_SIGMA" "EEG_BETA"
FILTER command to apply a different band-pass filter to each of the channels:
leval( "FILTER sig=EEG_DELTA bandpass=0.5,4 tw=1 ripple=0.02" )
leval( "FILTER sig=EEG_THETA bandpass=4,8 tw=1 ripple=0.02" )
leval( "FILTER sig=EEG_ALPHA bandpass=8,12 tw=1 ripple=0.02" )
leval( "FILTER sig=EEG_SIGMA bandpass=12,15 tw=1 ripple=0.02" )
leval( "FILTER sig=EEG_BETA bandpass=15,30 tw=1 ripple=0.02" )
We then epoch the data using lepoch():
ne <- lepoch()
nsrr02 : 6 signals, 10 annotations, 09:57:30 duration, 1195 unmasked 30-sec epochs, and 0 masked
We can use the ldata() function to pull out epochs of data for all 6 signals (i.e. as specified by giving lchs() as the second argument), e.g. here for epoch 135:
d <- ldata( 135 , lchs() )
d:
> head(d)
INT E SEC EEG EEG_DELTA EEG_THETA EEG_ALPHA
1 4020.00->4050.00 135 4020.000 -0.4901961 -3.473108 -0.06260304 3.4154174
2 4020.00->4050.00 135 4020.008 -2.4509804 -3.452390 -0.53536473 2.2660953
3 4020.00->4050.00 135 4020.016 -4.4117647 -3.307357 -1.13706143 0.4709635
4 4020.00->4050.00 135 4020.024 -5.3921569 -3.051824 -1.76561958 -1.5923434
5 4020.00->4050.00 135 4020.032 -6.3725490 -2.678883 -2.30284877 -3.4422047
6 4020.00->4050.00 135 4020.040 -7.3529412 -2.188535 -2.65204775 -4.5915269
EEG_SIGMA EEG_BETA
1 -1.9357408 -0.2801375
2 -2.7737269 1.0227155
3 -2.3793805 1.6093081
4 -0.9104402 0.8806983
5 0.9774931 -0.6320931
6 2.4513626 -1.3668775
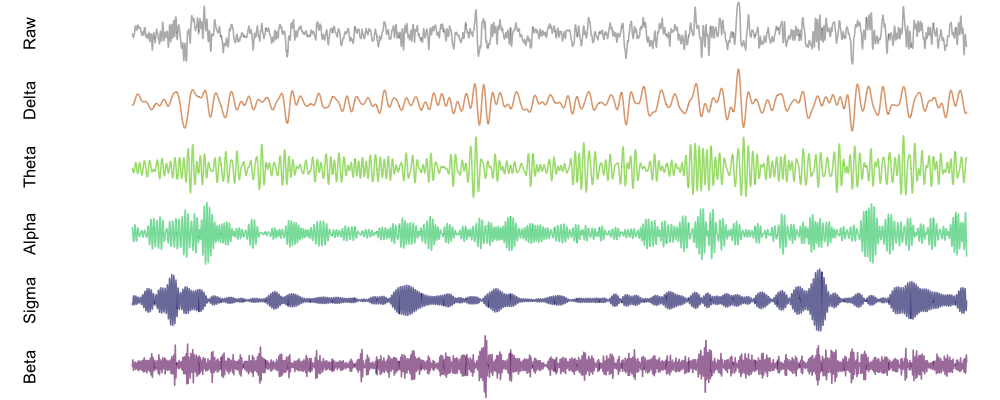
We can plot a data frame in this format with a R's internal functions, such as:
f1 <- function(d) {
par(mfcol=c(6,1),mar=c(0,5,0,0))
plot( d$EEG , type="l" , col=rgb(50,50,50,100,max=255) , axes=F , ylab="Raw" )
plot( d$EEG_DELTA , type="l" , col=rgb(200,100,00,150,max=255) , axes=F , ylab="Delta" )
plot( d$EEG_THETA , type="l" , col=rgb(100,200,0,150,max=255) , axes=F , ylab="Theta" )
plot( d$EEG_ALPHA , type="l" , col=rgb(0,200,100,150,max=255) , axes=F , ylab="Alpha" )
plot( d$EEG_SIGMA , type="l" , col=rgb(0,0,100,150,max=255) , axes=F , ylab="Sigma" )
plot( d$EEG_BETA , type="l" , col=rgb(100,0,100,150,max=255) , axes=F , ylab="Beta" )
}
Then, running f1( d ) will produce this plot (although, note that different
signals are not uniformly scaled in this particular representation):
FILTER-DESIGN
Frequency and impulse responses for FIR filters designed via the Kaiser window method
Filters specified by the FILTER command use the Kaiser
window
method to design the filter. The FILTER-DESIGN command (or Luna
--fir option) can be used to show the properties of these filters.
This command does not depend on any EDFs to be present, and so can be run without a sample-list or EDF (see the example below).
Parameters
| Parameter | Example | Description |
|---|---|---|
fs |
fs=256 |
Sampling rate |
bandpass |
bandpass=0.3,35 |
Band-pass filter between 0.3 and 35 Hz |
lowpass |
lowpass=35 |
Low-pass filter with cutoff of 35 Hz |
highpass |
highpass=0.3 |
High-pass filter with cutoff of 0.3 Hz |
bandstop |
bandstop=55,65 |
Band-stop filter between 0.3 and 35 Hz |
The FIR design design approaches are as for the FILTER command: either through the window method
(with either a Kaiser window -- tw and ripple -- or fixing the FIR order) or reading from a file:
| Parameter | Description |
|---|---|
ripple |
Ripple (as a proportion) |
tw |
Transition width (in Hz) |
file |
Read FIR coefficients from a file |
order |
Fix FIR order |
rectangular |
Specify a rectangular window |
bartlett |
Specify a Bartlett window |
hann |
Specify a Hann window |
blackman |
Specify a Blackman window |
Output
Per-filter basics (strata: none)
| Variable | Description |
|---|---|
FIR |
Label for FIR filter (constructed from input parameters) |
FS |
Sampling rate (from fs input parameter) |
NTAPS |
Filter order (number of taps) |
Filter coefficients (strata: F x TAP)
| Variable | Description |
|---|---|
TAP |
Tap |
W |
Filter coefficient |
Frequency response characteristics (strata: F x FIR)
| Variable | Description |
|---|---|
F |
Frequency (Hz) |
FIR |
FIR filter label |
MAG |
Magnitude |
MAG_DB |
Magnitude (dB) |
PHASE |
Phase |
Impulse response (strata: FIR x SEC)
| Variable | Description |
|---|---|
F |
Time (seconds) |
FIR |
FIR filter label |
IR |
Impulse response |
Example
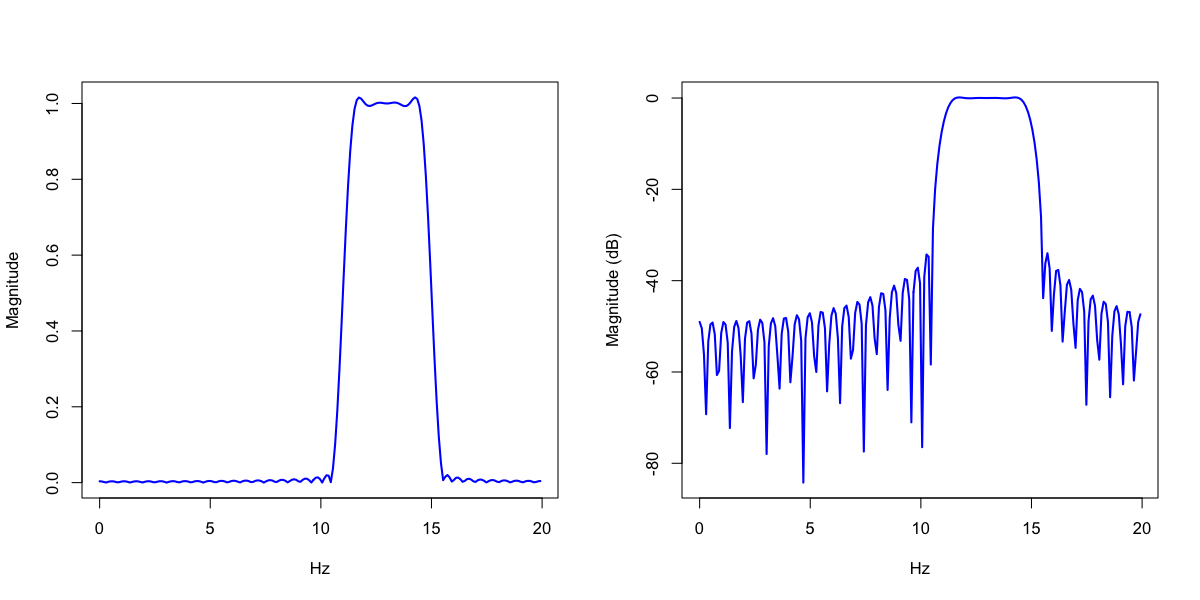
Consider a band-pass filter in the sigma band, 11 to 15Hz, applied to a signal with 200 Hz sampling rate. Transition frequencies 11Hz and 15Hz have -6dB attenuation (i.e. 50% amplitude ratio).
If specifying the --fir option, Luna expects the parameters above
via standard input, e.g. piped from the echo shell command, with the
output still sent to a database out.db:
echo "fs=200 bandpass=11,15 ripple=0.02 tw=1" | luna --fir -o out.db
Examining the out.db, we see that this command produces the following output:
destrat out.db
--------------------------------------------------------------------------------
out.db: 1 command(s), 1 individual(s), 7 variable(s), 4207 values
--------------------------------------------------------------------------------
command #1: c1 Mon Feb 25 16:11:40 2019 FIR-DESIGN
--------------------------------------------------------------------------------
distinct strata group(s):
commands : factors : levels : variables
----------------:-------------------:---------------:---------------------------
[FIR-DESIGN] : FIR : 1 level(s) : FS NTAPS
: : :
[FIR-DESIGN] : F FIR : 1025 level(s) : MAG MAG_DB PHASE
: : :
[FIR-DESIGN] : FIR TAP : 365 level(s) : W
: : :
[FIR-DESIGN] : FIR SEC : 765 level(s) : IR
: : :
----------------:-------------------:---------------:---------------------------
The first stratum (FIR) gives the sampling rate (FS) as input, and
the derived order of the filter (NTAPS):
destrat out.db +FIR-DESIGN -r FIR
ID FIR FS NTAPS
. BANDPASS_11..15_0.02_1 200 365
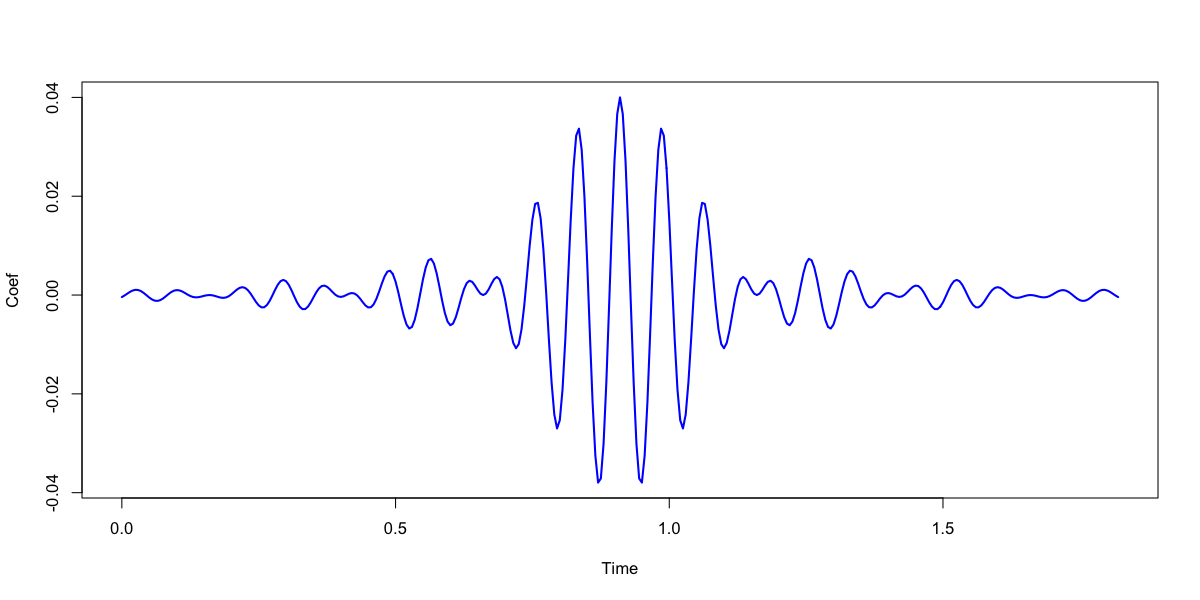
The actual filter coefficients are in the stratum defined by FIR x TAP:
destrat out.db +FIR-DESIGN -r FIR TAP > fir.txt
In R, we can plot the filter coefficients:
w <- read.table("fir.txt",header=T)
plot( w$TAP/200 , w$W , type="l", lwd=2, col="blue", ylab="Coef", xlab="Time" )
The amplitude (magnitude) response of the filter is in the F x FIR stratum:
destrat out.db +FIR-DESIGN -r F FIR > fir2.txt
d <- read.table("fir2.txt", header=T)
d <- d[ d$F < 20 , ]
par(mfcol=c(1,2))
plot(d$F, d$MAG , type="l",lwd=2,col="blue", xlab="Hz",ylab="Magnitude")
plot(d$F, d$MAG_DB, type="l",lwd=2,col="blue", xlab="Hz",ylab="Magnitude (dB)")