Merging and converting EDFs
In this vignette, we illustrate how to use Luna to merge multiple EDFs
into a single EDF including any associated annotations, using the --merge
and --bind commands.
This procedure allows for gaps between component EDFs, in which case a
discontinuous EDF+D file will be emitted. The second part of this vignette shows
how to convert an EDF+D (whether from --merge or any other source) to a standard EDF,
once again appropriately handling any associated annotations.
Merging multiple EDFs
Luna's --merge command can concatenate two or more EDFs, under
certain conditions:
- all concatenated EDFs must have similar EDF headers: i.e. the same number and order of channels and sample rates
- the physical/digital min/max values can be different - these will be rescaled in the final merged EDF
- EDFs can have gaps between them, but they are not allowed to overlap in time
- currently, only standard EDFs (not EDF+) files can be concatenated
The basic use of this command, here to join three EDFs, is as follows:
luna --merge file1.edf file2.edf file3.edf
which will generate a new EDF called (by default) merged.edf. To explicitly specify an EDF filename (and Patient ID too),
add edf and `id arguments: to save as f001.edf, use the following:
luna --merge edf=f001.edf id=id001 file1.edf file2.edf file3.edf
The merge command can run in two modes: one in which EDF start times
are effectively ignored: all EDFs are directly concatenated in the
order as given on the command line (the resulting merged EDF will have
a start time/date based on the earliest EDF. This mode (which is
enabled by adding the argument fixed=T to the options of --merge)
will always generate a standard EDF file (i.e. no gaps) but will
potentially lose information about the timing between EDFs.
In contrast, the default mode for --merge is to examine the start
dates/times of all EDFs, to check that they do not overlap, and then
assemble them into a single EDF. If gaps are implied between EDFs,
the resulting merged EDF will be an EDF+D (i.e to represent the
discontinuities between input EDFs). Otherwise, a standard EDF will be
emitted as above.
Example
To generate a trivial example for merging two EDFs, for convenience we'll take a single (whole-night) EDF and simply try concatenating
it to itself, i.e. to generate a file that is two nights' worth of data stacked into one. Consider a starting EDF called f.edf, which contains only two EEG channels in this contrived example:
luna f.edf -s DESC
EDF filename : f.edf
ID : f
Clock time : 20.53.59 - 08.07.29
Duration : 11:13:30 40410 sec
Signals : C3[128] C4[128]
If we initially attempt to merge it with itself as is, Luna will correctly complain:
luna --merge f.edf f.edf
attached 2 EDFs
writing merged data:
ID : merged1
EDF filename : merged.edf
------------------------------------------------------------
extracting start times from EDFs --> seconds since 1/1/85 00:00
EDF 0 date: 01.01.85 time: 20.53.59 days: 0 --> secs:75239
EDF 1 date: 01.01.85 time: 20.53.59 days: 0 --> secs:75239
error : EDFs with identical start times specified: 01.01.85 20.53.59
As noted above, we can use the fixed=T option to ignore start times
of EDFs – in which case Luna will join the EDFs in the order specified
after the -—merge (although, in this contrived example, these are
identical of course):
luna --merge fixed=T f.edf f.edf
attached 2 EDFs
writing merged data:
ID : merged1
EDF filename : merged.edf
good, all EDFs have merge-compatible headers
expecting 80820 records (each of 1 sec) in the new EDF
adding timeline
adding 80820 empty records...
compiling channel C3
compiling channel C4
writing merged EDF as merged.edf
data are not truly discontinuous
writing as a standard EDF
writing 2 channels
saved new EDF, merged.edf
Aligned EDFs
For a more realistic example, we'll create two EDFs (both from
f.edf) with different start dates and times, but that are aligned in
the sense that the second follows directly after the first ends. As
noted above, the original EDF had a start time of 20.53.59 and was
40,410 seconds in duration. We will generate one EDF with the same
start time, but we'll also explicitly set a non-null start date too,
using SET-HEADERS followed by WRITE to make a new EDF:
luna f.edf -s 'SET-HEADERS start-date=22.02.22 start-time=20.53.59 & WRITE edf=pt1'
pt1.edf:
luna pt1.edf -s DESC
Clock time : 20.53.59 - 08.07.29
Duration : 11:13:30 40410 sec
Next, we'll generate a second EDF that starts exactly after the first one ends, i.e. 08.07.29.
(Note that the end time given by DESC is one time point past the end
of the interval, consistent with how Luna expects intervals for
annotations, i.e. meaning up until but not including that time. That
is, the last record starts at 08.07.29 and is 1 second long.)
luna f.edf -s 'SET-HEADERS start-date=23.02.22 start-time=08.07.29 & WRITE edf=pt2'
luna --merge pt1.edf pt2.edf
attached 2 EDFs
writing merged data:
ID : merged1
EDF filename : merged.edf
------------------------------------------------------------
extracting start times from EDFs --> seconds since 1/1/85 00:00
EDF 0 date: 22.02.22 time: 20.53.59 days: 13566 --> secs:1172177639
EDF 1 date: 23.02.22 time: 08.07.29 days: 13567 --> secs:1172218049
------------------------------------------------------------
ordered EDFs (seconds past 1/1/1985 00:00:00)
ordered EDFs : prev start = 1172177639 ; this start = 1172218049
implied duration of previous based on # records = 40410
implied duration of previous based on current EDF start = 40410
implies exactly contiguous EDFs
no gaps found between EDFs - will generate an EDF (or EDF+C)
good, all EDFs have merge-compatible headers
expecting 80820 records (each of 1 sec) in the new EDF
adding timeline
adding 80820 empty records...
compiling channel C3
compiling channel C4
writing merged EDF as merged.edf
data are not truly discontinuous
writing as a standard EDF
writing 2 channels
saved new EDF, merged.edf
That is, Luna can see that the resulting concatenated EDF is not
actually discontinuous (as the stop of pt1.edf and the start of
pt2.edf are exactly aligned) and so saves it as a standard EDF. The
duration is now 80,820 seconds, i.e. twice 40,410 seconds,
the duration of each original EDF.
luna merged.edf -s DESC
EDF filename : merged.edf
ID : merged
Clock time : 20.53.59 - 19.20.59
Duration : 22:27:00 80820 sec
# signals : 2
Signals : C3[128] C4[128]
Overlapping EDFs
Here we specify some overlap between the two EDFs and attempt to merge: we'll set the
start time of pt2.edf to be 6am, i.e. about two hours before pt1.edf actually ends:
luna f.edf -s 'SET-HEADERS start-date=23.02.22 start-time=06.00.00 & WRITE edf=pt2'
If we now try to merge:
luna --merge pt1.edf pt2.edf
attached 2 EDFs
writing merged data:
ID : merged1
EDF filename : merged.edf
------------------------------------------------------------
extracting start times from EDFs --> seconds since 1/1/85 00:00
EDF 0 date: 22.02.22 time: 20.53.59 days: 13566 --> secs:1172177639
EDF 1 date: 23.02.22 time: 06.00.00 days: 13567 --> secs:1172210400
------------------------------------------------------------
ordered EDFs (seconds past 1/1/1985 00:00:00)
ordered EDFs : prev start = 1172177639 ; this start = 1172210400
implied duration of previous based on # records = 32761
implied duration of previous based on current EDF start = 40410
*** warning -- overlapping EDFs implied
error : found overlapping EDFs -- bailing, cannot merge
That is, Luna correctly points to the issue and refuses to do anything further (after giving some output that shows the implied overlap).
Info
Internally, Luna uses seconds past midnight at 1/1/1985 to
compare EDF start date/time combinations, i.e. this is what
secs:1172177639 above refers to.
Gapped EDFs
Third, we will introduce a gap between recordings, as might typically occur if a
recording is paused for some reason. Here, we will set the second EDF
to start an hour or so after the first one ends (09:15:00 instead of
08:07:29):
luna f.edf -s 'SET-HEADERS start-date=23.02.22 start-time=09.15.00 & WRITE edf=pt2'
Now the merge will generate an EDF+D with a gap:
luna --merge pt1.edf pt2.edf
attached 2 EDFs
writing merged data:
ID : merged1
EDF filename : merged.edf
------------------------------------------------------------
extracting start times from EDFs --> seconds since 1/1/85 00:00
EDF 0 date: 22.02.22 time: 20.53.59 days: 13566 --> secs:1172177639
EDF 1 date: 23.02.22 time: 09.15.00 days: 13567 --> secs:1172222100
------------------------------------------------------------
ordered EDFs (seconds past 1/1/1985 00:00:00)
ordered EDFs : prev start = 1172177639 ; this start = 1172222100
implied duration of previous based on # records = 44461
implied duration of previous based on current EDF start = 40410
implies gap between previous and current - will output an EDF+D
found gaps between EDFs - will generate an EDF+D
good, all EDFs have merge-compatible headers
expecting 80820 records (each of 1 sec) in the new EDF
adding timeline
adding 80820 empty records...
compiling channel C3
compiling channel C4
set EDF+D timeline for 80820 records
writing merged EDF as merged.edf
data are truly discontinuous
writing 3 channels
saved new EDF+D, merged.edf
That is, the emitted EDF+D file contains an EDF Annotations channel that specifies the start time of each EDF record
relative to the header start time. Now when we look at the DESC, Luna will give some extra information describing the
EDF+D features:
luna merged.edf -s DESC
EDF filename : merged.edf
ID : merged
Header start time : 20.53.59
Last observed time: 20.28.30
Duration : 22:27:00 80820 sec
Duration (w/ gaps): 23.34.31 84871 sec
# signals : 2
# EDF annotations : 1
Signals : C3[128] C4[128]
That is, although the duration of observed records is still 80,820
seconds, the total time spanned (i.e. including gaps) is longer
(84,871 seconds). We can also look at the structure of a discontinuous EDF+D more
explicitly with the SEGMENTS
command:
luna merged.edf -o out.db -s SEGMENTS
destrat out.db +SEGMENTS
ID NGAPS NSEGS
merged 1 2
destrat out.db +SEGMENTS -r SEG
ID SEG DUR_HR DUR_MIN DUR_SEC START START_HMS STOP STOP_HMS
merged 1 11.225 673.5 40410 0 20.53.59 40410 08.07.29
merged 2 11.225 673.5 40410 44461 09.15.00 84871 20.28.30
08.07.29 and 09.15.00:
destrat out.db +SEGMENTS -r GAP
ID GAP DUR_HR DUR_MIN DUR_SEC START START_HMS STOP STOP_HMS
merged 1 1.12527 67.5167 4051 40410 08.07.29 44461 09.15.00
Alternatively, we can use the Moonlight viewer (public test version
available at http://remnrem.net) to view the structure of an
EDF+D. After uploading merged.edf to this tool, the
Strucutre/Segments tab shows:
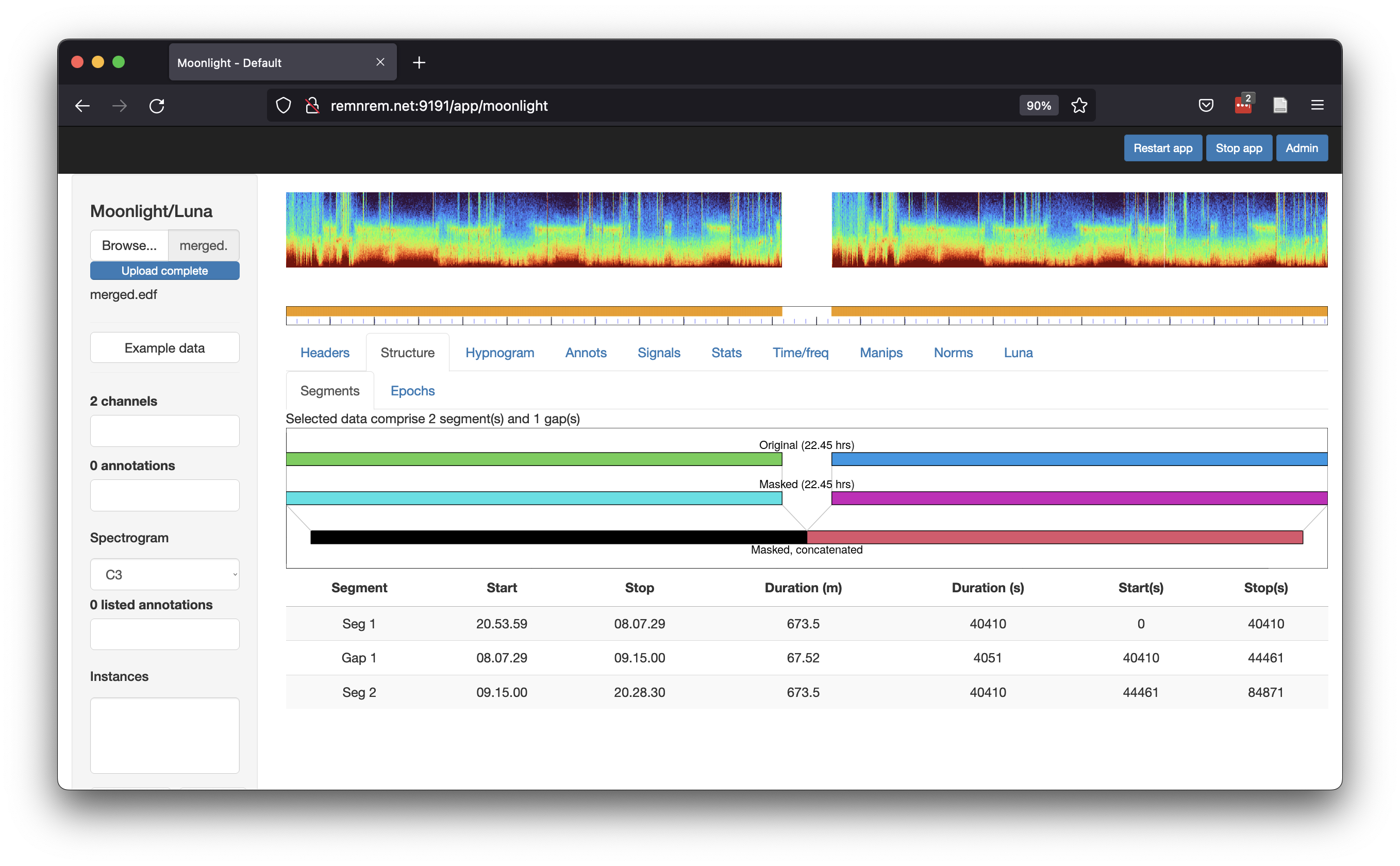
Note how the spectrogram is broken in the middle, indicating the gap between the two segments; also note how the first and second segments have identical spectrograms, as expected in this contrived example.
Merging annotations
The --merge command only concatenates EDF signal data. Typically,
we'll have associated annotation files (e.g. manual sleep staging)
that also require combining. Particularly if using Luna .annot format,
this is a trivial process, although some practical considerations still apply, as described here.
Although .annot files can be simply combined (e.g. via the command
line cat utility), if annotations files specify event times relative to
an EDF start (i.e. in elapsed seconds), these will need to be
adjusted prior to merging. Indeed, in our example we also have an XML
annotation file (f.xml) that contains sleep stage
annotations, with the intervals encoded as elapsed seconds from the
start of f.edf (i.e. seconds after 20.53.59).
Following from the above example, we can copy f.xml to produce two sets of annotations:
cp f.xml pt1.xml
cp f.xml pt2.xml
The most direct way to handle merging annotations is to use absolute
clock-time rather than elapsed time in each annotation file. (An
alternative not considered here is to manually add in the second
offsets, which is why --merge actually generates these offsets for
each file and outputs them.) We can generate clock-based
annotations using Luna's WRITE-ANONTS command, to generate (in this
case) two .annot files that have hh:mm:ss times. For the first
one:
luna pt1.edf annot-file=pt1.xml -s WRITE-ANNOTS file=pt1.annot minimal hms
For example, the first N1 epoch now in pt1.annot reads as
follows:
N1 . . 21:40:59 21:41:59 .
starting at 21:40:59 and lasting one minute (two epochs) until
21:41:59. In contrast, if exported without the hms flag, the same
line would have read (which mirrors how the event is stored in the original
EDF):
N1 . . 2820.000 2880.000 .
i.e. 2820 seconds (2820/60=47 minutes) after 20.53.59 is 21:40:59.
From v0.28, Luna also supports explicit date-time strings in
annotation files, which can be helpful to reduce ambiguity in longer
recordings that use clock-time (including multi-day/night recordings).
If we add dhms instead of hms, then WRITE-ANNOTS would instead
output this for this particular line:
N1 . . 22-2-2022-21:40:59 22-2-2022-21:41:59 .
i.e. in the form dd-mm-yyyy-hh:mm:ss. For this particular example -
as the merged EDF is almost 24 hours long - we will use this explicit
date-time representation, which should always be safer. (Note: in the
context of anonymized EDFs with dates set to 1/1/85 as per the EDF spec, for
multi-day recordings, we suggest to start with 2/1/85 instead, which as a non-null date value
will allow dates to be specified in annotation files, and will roll forwards as expected, 3/1/85, etc.)
Running the same process for the second EDF:
luna pt2.edf annot-file=pt2.xml -s WRITE-ANNOTS file=pt2.annot minimal dhms
pt2.annot which has the following for the corresponding line:
N1 . . 23-2-2022-10:02:00 23-2-2022-10:03:00 .
as 10:02:00 is 47 minutes after the 09:15 start time of pt2. That is,
although pt1.xml and pt2.xml were identical files, the newly
encoded .annot files explicitly reflect dates and times from the two EDFs,
and so can now be merged without loss of information or introducing ambiguity.
Note that we also specified the minimal option for WRITE-ANNOTS - this skips the header rows, which
makes it easier to combine the .annot files with a single cat command:
cat pt1.annot pt2.annot > merged.annot
Note that we do not even have to combine the annotation files, i.e. we can specify them individuals (on the command line, or in a sample list):
luna merged.edf annot-file=pt1.annot,pt2.annot ...
If we upload both merged.edf and merged.annot to Moonlight, we can now see the annotations are appropriately aligned
with the signal data (i.e. see the extended hypnogram is now added beneath the spectrogram):
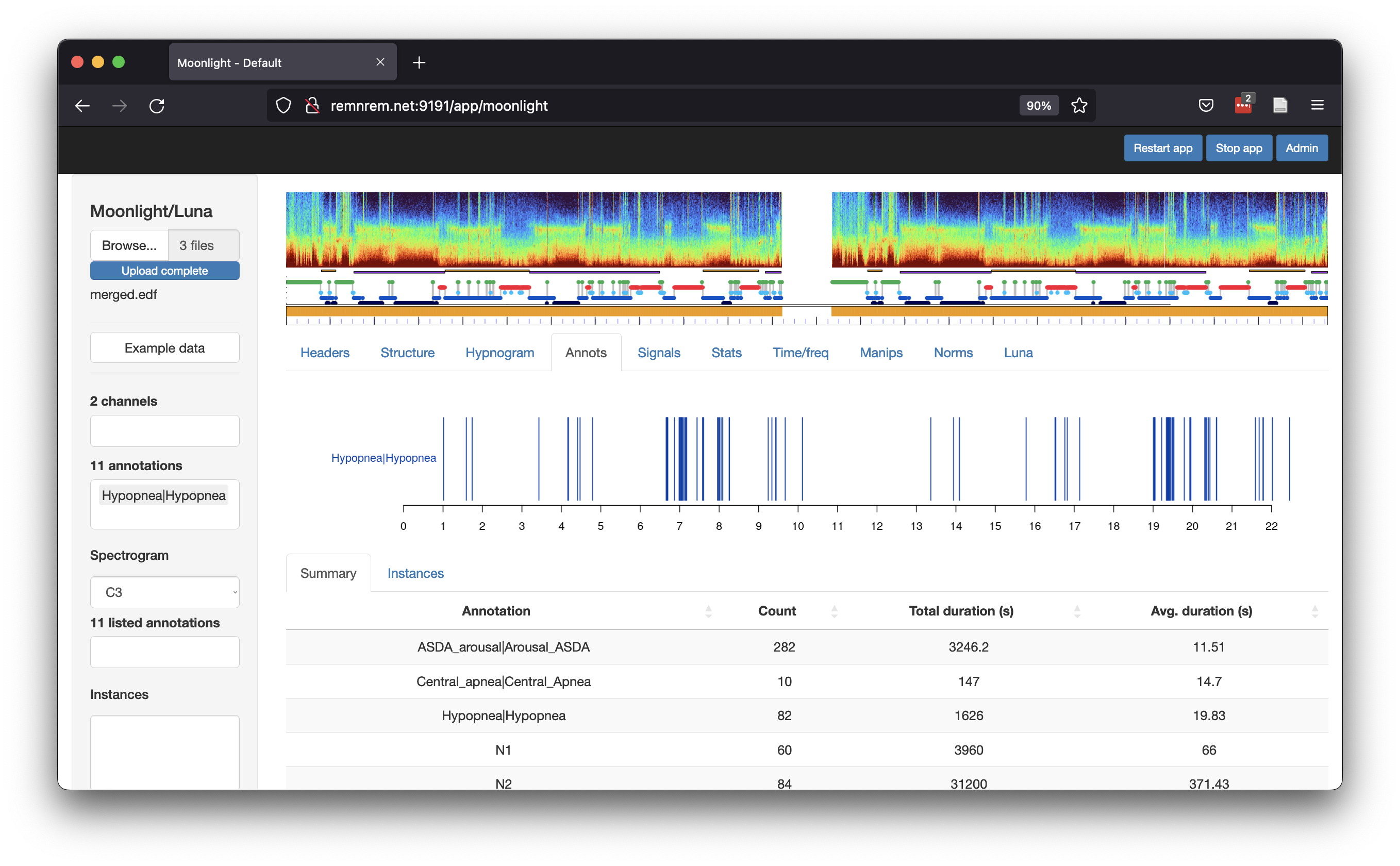
Again - in this contrived example we have the unusual situation of duplicated, identical whole-night recordings.
Typically, --merge would expect single-night recordings but where the EDFs are fragmented across two or more files, e.g.
due to pauses in the recording, or idiosyncrasies of the EDF export process, etc. Still, the principles are the same from
--merge's perspective.
EDF+D to EDF conversion
In the second part of this vignette, we consider how to work with EDF+D files - that is, in the context of sleep recordings, this typically means recordings that contain gaps.
Note that this older vignette also contains information on working with EDF+D files. The primary focus of this section is how to convert an EDF+D to a standard (continuous) EDF file. In many cases, standard EDFs can be easier to work with. Indeed, some tools might not accept EDF+(D) files at all. Below we a) use Luna to artificially generate a toy EDF+D dataset, and then b) use Luna to convert it back to a standard EDF, i.e. splicing out the gaps. Although the principal conversion is straightforward, the primary issue covered here is how to keep annotations aligned with transformed files.
Simulating data
Here we use the simple SIGGEN command to simulate a 30-second dataset with
three simple pulses, each lasting 50 samples (0.5 seconds, given a sample rate of 100 Hz):
luna . --nr=30 --rs=1 \
-s ' SIGGEN sig=S1 sr=100 impulse=0.1,1,50,0.6,1,50,0.9,1,50
& S2A sig=S1 encoding=A,1
& WRITE edf=standard
& WRITE-ANNOTS file=standard.annot '
That is, the . indicates an empty EDF
of 30 records, each 1 second long; we then generate a signal called
S1 with 100 Hz sample rate, and pulses (values of 1 versus 0)
starting at 10%, 60% and 90% of the recording (i.e. 3 seconds, 18
seconds and 27 seconds from the EDF start).
We next add an annotation that corresponds to these pulses using the
S2A command, called A. Finally, we
write the signal data to a standard EDF file (called standard.edf)
and the annotation to a file called standard.annot. The upshot of
this is visually depicted here, i.e. plotting data in the file f.0
generated by the MATRIX command:
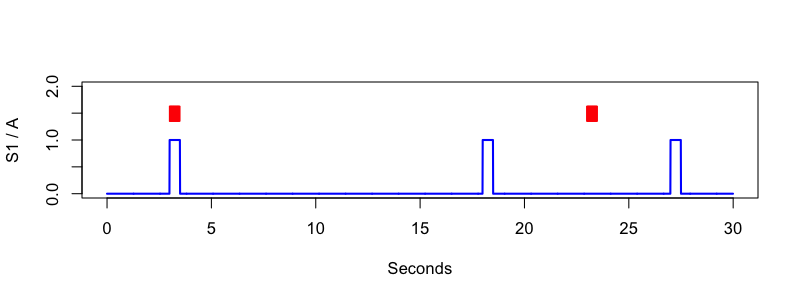
luna standard.edf annot-file=standard.annot -s 'MATRIX sig=S1 annot=A file=f.0 '
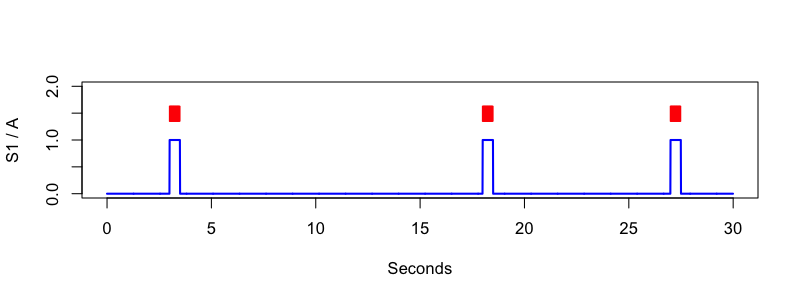
Making an EDF+D
So far, we have generated a standard EDF (30 seconds in duration) and
a simple annotation file, with three events corresponding to the
pulses in the signal S1. The next step is to make this an EDF+D
with gaps (i.e. so that we can show the process of converting it back
again to standard EDF). For this, we can use Luna's helper command SET-TIMESTAMPS
to explicitly assign the offset of each EDF record, taking data from a file. Implicitly,
the standard EDF has records that start at 0, 1, 2, ... seconds past the EDF start. We
will generate a similar explicit file, but with two gaps insert (an extra 5 seconds between the 16th
and 17th records using base-0 counting, and an extra 7.4525 seconds between the 26th and 27th records). Although we use awk here,
one could simply type out the numbers generated below instead:
echo | awk ' BEGIN { s = 0 }
{ for (i=0; i<30; i++)
{ print s ; s += 1 ;
if ( i == 15 ) s += 5 ;
if ( i == 25 ) s += 7.4525 } } ' > ts.txt
The contents of ts.txt then has 30 rows, which specify the new EDF start times for the records in standard.edf:
0
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
21
22
23
24
25
26
27
28
29
30
38.4525
39.4525
40.4525
41.4525
Here we apply those new times and write a new EDF
luna standard.edf -s ' SET-TIMESTAMPS file=ts.txt & WRITE edf=plus '
SET-TIMESTAMPS we see the following:
read 30 timestamps
set EDF+D timeline for 30 records
updated EDF+D time-track
WRITE:
data are truly discontinuous
writing 2 channels
saved new EDF+D, plus.edf
plus.edf
is as expected:
luna plus.edf -s ' DESC & RECS '
EDF filename : plus.edf
ID : plus
Header start time : 00.00.00
Last observed time: 00.00.42
Duration : 00:00:30 30 sec
Duration (w/ gaps): 00.00.42 42.4525 sec
# signals : 1
# EDF annotations : 1
Signals : S1[100]
That is, we have 30 seconds of data (30 1 second records) and an extra 5+7.4525 = 12.4525 seconds of gaps, leading to the total duration of 42.4525 seconds.
The RECS command dumps the EDF record structure/times to the console
- we see a pattern of times that mirrors those specified in ts.txt
(some columns omitted here for clarity, and the two empty rows inserted manually, to show
the gaps):
Record Time
1 0.0000->1.0000
2 1.0000->2.0000
3 2.0000->3.0000
4 3.0000->4.0000
5 4.0000->5.0000
6 5.0000->6.0000
7 6.0000->7.0000
8 7.0000->8.0000
9 8.0000->9.0000
10 9.0000->10.0000
11 10.0000->11.0000
12 11.0000->12.0000
13 12.0000->13.0000
14 13.0000->14.0000
15 14.0000->15.0000
16 15.0000->16.0000
17 21.0000->22.0000
18 22.0000->23.0000
19 23.0000->24.0000
20 24.0000->25.0000
21 25.0000->26.0000
22 26.0000->27.0000
23 27.0000->28.0000
24 28.0000->29.0000
25 29.0000->30.0000
26 30.0000->31.0000
27 38.4525->39.4525
28 39.4525->40.4525
29 40.4525->41.4525
30 41.4525->42.4525
Note
Note, instead of using SET-TIMESTAMPS we could have simply
generated an EDF+D file using a mask and restructuring an
in-memory dataset, as after any restucturing Luna's in-memory EDF
is effectively an EDF+D, i.e. in that it explicitly represents
gaps by tracking the start time of each EDF record explicitly.
For example (noting that we need to set a smaller epoch size here,
given the total recording length for this toy dataset is only 30
seconds:
luna standard.edf -s ' EPOCH dur=1 & MASK epoch=1-10,15-30 & RE & WRITE edf=plus '
Finally, we can also alter the annotations originally created,
cat standard.annot
class instance channel start stop meta
A . S1 3.000 3.500 .
A . S1 18.000 18.500 .
A . S1 27.000 27.500 .
Here we add in gaps between the pulse events that mirror the gaps inserted above in the signal.
awk ' NR < 3 { print $0 }
NR == 3 { print $1,$2,$3,$4+5,$5+5,$6 }
NR == 4 { print $1,$2,$3,$4+12.4525,$5+12.4525,$6 } ' OFS="\t" standard.annot > plus.annot
cat plus.annot
class instance channel start stop meta
A . S1 3.000 3.500 .
A . S1 23 23.5 .
A . S1 39.4525 39.9525 .
The details of the above are not important: the bottom line of all this is that we now have an EDF+D with gaps, and some annotations that are temporally aligned with the signal data, as shown here:
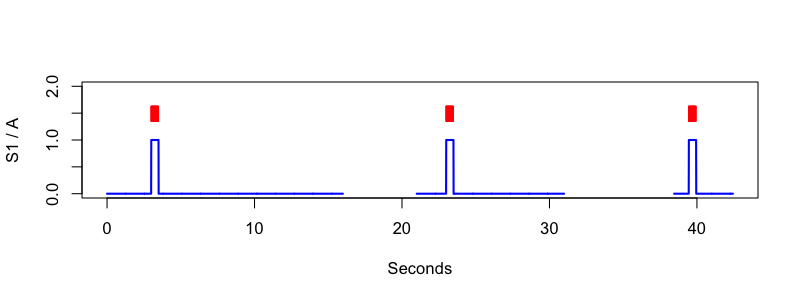
Conversion back to EDF
Now, force the EDF+D to a standard EDF
luna plus.edf -s 'WRITE edf=standard2 force-edf '
EDF filename : standard2.edf
ID : standard2
Clock time : 00.00.00 - 00.00.29
Duration : 00:00:30 30 sec
# signals : 1
Signals : S1[100]
luna standard.edf annot-file=plus.annot -s 'EPOCH dur=1 & MATRIX sig=S1 annot=A file=f.2 '
To get the annotations to align, we must add the collapse flag to WRITE-ANNOTS
luna plus.edf annot-file=plus.annot -s 'WRITE-ANNOTS collapse file=standard2.annot '
cat standard2.annot
class instance channel start stop meta
A . S1 3.000 3.500 .
A . S1 18.000 18.500 .
A . S1 27.000 27.500 .
luna standard2.edf annot-file=standard2.annot -s 'MATRIX sig=S1 annot=A file=f.3 '
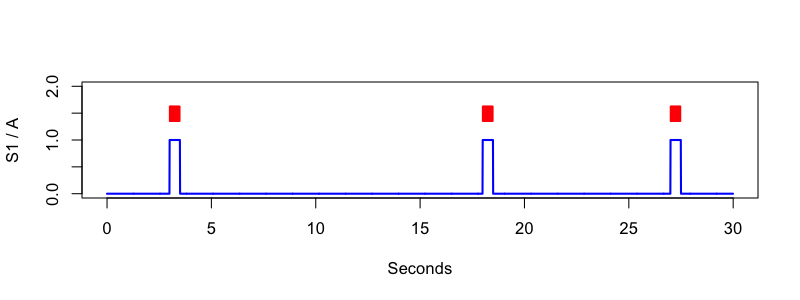
Finally, it is also possible to use the SEGMENTS command with an EDF+D (whether a true EDF+D file, or
an in-memory EDF+D, i.e constructed after setting masks and calling RESTRUCTURE) to generate new annotations
that reflect the segments (and gaps) in an EDF+D file:
luna plus.edf -o out.db -s ' EPOCH dur=1 & SEGMENTS annot & ANNOTS '
Hint
As above, some commands implicitly use epochs to iterate over data, including the ANNOTS command. Because we are working with
an artificially short recording, that does not have any individual segments greater than 30 seconds in length, it is necessary
to set a shorter epoch length (here 1 second) so that those commands produce any output
destrat out.db +ANNOTS -r ANNOT INST T -v START STOP
ID ANNOT INST T START STOP
plus segment 1 0_16000000000 0 16
plus gap 1 16000000000_21000000001 16 21.000000001
plus segment 2 21000000000_31000000000 21 31
plus gap 2 31000000000_38452500001 31 38.452500001
plus segment 3 38452500000_42452500000 38.4525 42.4525
Note that gap annotations are actually expanded - one time-point past the end
of the gap (1e-09 seconds). This is done purposefully, so that we used with
WRITE-ANNOTS collapse, the gap annotations are not entirely spliced out,
but rather form single annotations at the end and start of the respective flanking segments,
as above. Also note that the instance ID (INST) is set to the
number of the segment/gap.
Bottom line
Wrapping this all together, starting from an EDF+D with annotations, in order to produce a) a standard EDF containing only contiguous EDF records, b) a shifted set of annotations, that are aligned with the spliced-out EDF signals, and c) a set of new annotations for the new standard EDF that show where there were discontinuities in the original EDF+D, we can do the following, single command:
luna plus.edf annot-file=plus.annot -s 'SEGMENTS annot & WRITE-ANNOTS collapse file=new.annot & WRITE force-edf edf=new'
cat new.annot
class instance channel start stop meta
segment 1 . 0.000 16.000 .
A . S1 3.000 3.500 .
gap 1 . 16.000 16.000 .
segment 2 . 16.000 26.000 .
A . S1 18.000 18.500 .
gap 2 . 26.000 26.000 .
segment 3 . 26.000 30.000 .
A . S1 27.000 27.500 .
We can now visualize the new dataset. This is optional, but we can make a new sample
list to group the EDF and annotation files, instead of using annot-file each time:
luna --build . | grep new.edf > s.lst
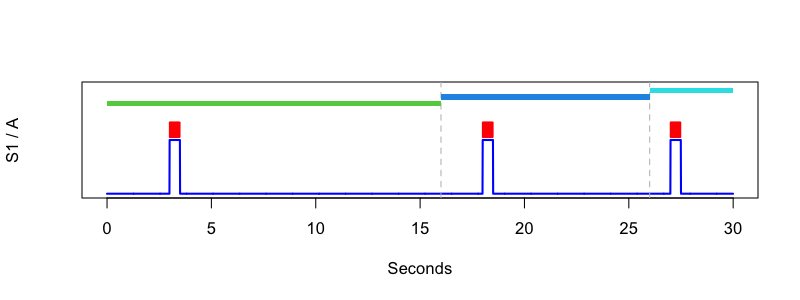
For reference, here was the R code to generate this figure:
lattach( lsl( "s.lst" ) , 1 )
d <- ldata( 1 , "S1" , "A" )
png( file = "edfplus-f4.png" , width=800 , height=300 , units="px" , res=100)
plot( d$SEC , d$S1 , type="l" , ylim=c(0,2) , lwd=2 , col="blue" , xlab = "Seconds" , ylab = "S1 / A" , yaxt='n' )
points( d$SEC[ d$A == 1 ] , d$A[ d$A == 1 ]+0.2 , type="p" , pch="|" , col = "red" )
abline( v = unique( unlist( lannots( "gap" ))) , lty=2, col="gray" )
segs <- lannots( "segment" )
for (s in 1:length(segs) ) {
seg <- segs[[s]]
rect( seg[1] , 1.5 + s/8 , seg[2] , 1.6+s/8 , col=s+2 ,border=NA)
}
dev.off()