Prediction models
Feature-based prediction models
The PREDICT command is designed to take a pre-existing linear
model with predictors (features) corresponding to metrics that Luna emits
(e.g. spectral power, etc). In conjunction
with a suitable script to estimate those metrics, PREDICT combines
model and data to make a prediction. Essentially, this framework aims
to provide a one-step procedure for going from (raw) PSG/EEG data as
input, to a model-based prediction as output.
For example, one PREDICT model supports the prediction of the
so-called brain age index using the NREM EEG, based on a model from
Sun et al (2019). This
model has also been incorporated into the Moonlight
viewer. Over time, different models as well as
support for model classes beyond linear models will be compiled here.
| Command | Description |
|---|---|
| Overview | Overview of the PREDICT framework |
PREDICT |
Make a prediction given a model and pre-calculated features |
| Models | Currently supported models |
Overview
The PREDICT command assumes the following components:
-
a model specification that defines a set of features, along with their weights and population means/standard deviations, used to construct a feature vector for each individual/EDF
-
a paired Luna script that generates the required features, using the
CACHE recordmechanism to pass those feature values to thePREDICTcommand -
optionally, a dataset of normalized values from the training dataset, to support kNN-based imputation of missing data
The primary workflow is as follows:
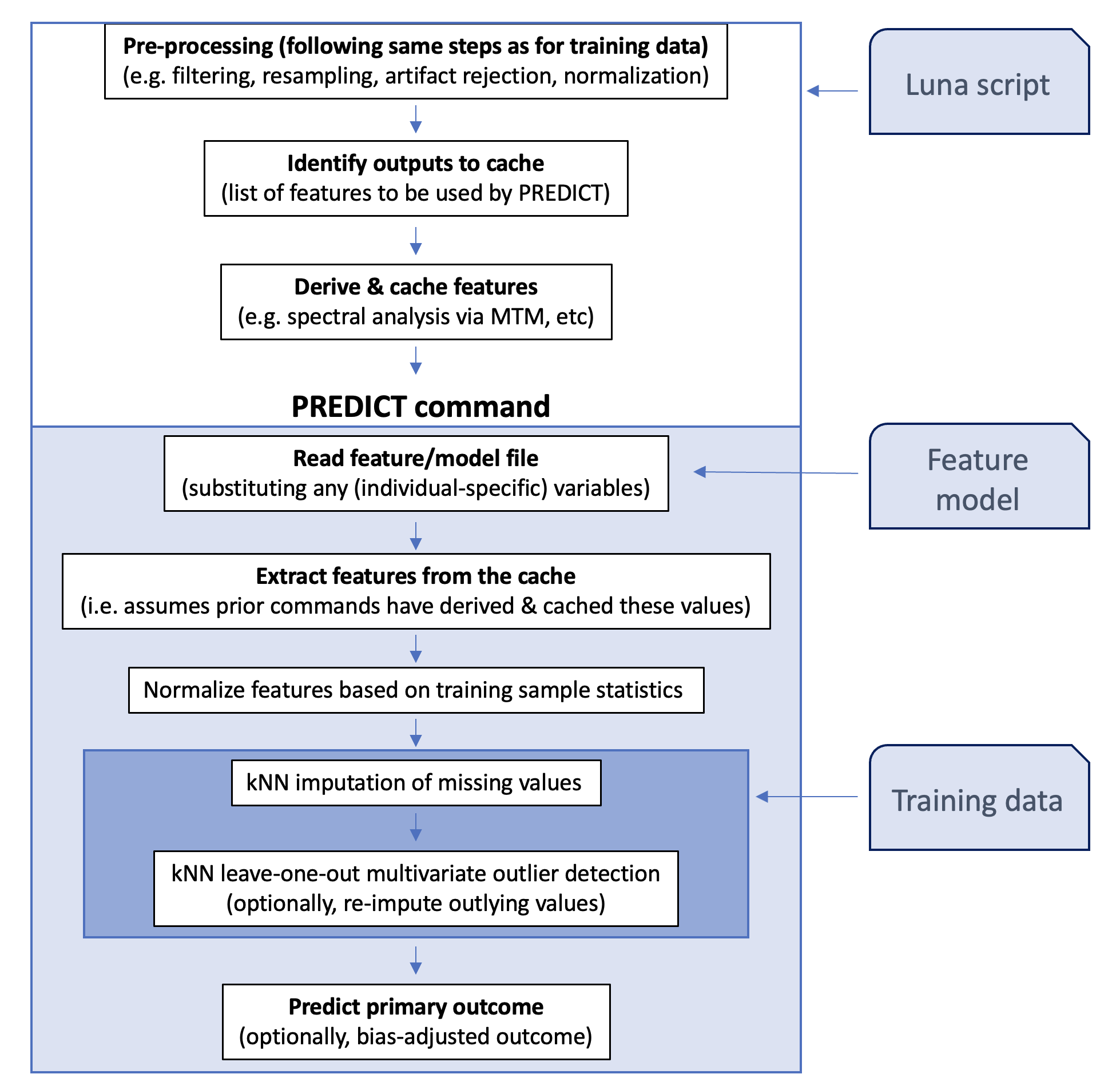
Info
As a downstream user of the PREDICT command (i.e. when using pre-defined models to make predictions)
most of the details on this page are unnecessary and you can skip ahead to the documentation on the main PREDICT
command itself.
The details below are for reference, aimed at individuals who want to use this framework to bring their own models into this framework.
Model specification file
A model specification file contains the following components:
-
feature definitions
-
definitions for special variables (e.g. model intercept), e.g.
Each feature definition has the following terms:
| Term | Example | Description |
|---|---|---|
| label | delta_theta_mean_C_N3 |
Arbitrary label for the feature |
CMD |
CMD=MTM |
Specifies the Luna command used to generate the feature |
VAR |
VAR=RATIO |
Specifies the exact Luna variable from that command |
STRATA |
STRATA=STG/N3,B1/DELTA,B2/THETA |
Specifies any additional strata |
CH |
CH=${cen} |
Specifies which channel(s) to consider |
m |
m=0.52 |
Population mean for this feature |
sd |
sd=0.23 |
Population standard deviation for this feature |
b |
b=-0.7 |
Coefficient for this feature (for a standardized metric) |
LOG |
LOG=1 |
Use the natural log of this feature, Z = sign(X) * log1p(|X|) |
REQ |
REQ=1 |
Set to 1/0 to indicate if a feature is required to be non-missing |
CHS |
CHS=C3+F3,C4+F4 |
Specifies pairwise channels, e.g. for metrics such as coherence (here two pairs: C3-F3 and C4-F4) |
DIR |
DIR=1 |
Set to 1/0 to indicate if a pairwise statistic is directional, i.e. if m(A,B) = -m(B,A) versus m(A,B) = m(B,A) |
VALUE |
VALUE=${x} |
Set a feature based on a variable rather than a cache value |
Typically, only the first eight terms will be used. The terms CHS
and DIR are only applicable for channel-pairwise metrics,
e.g. coherence. As a concrete example, this is one line (of 13 terms in the whole model)
from the Sun et al. model:
delta_theta_mean_C_N3
CMD=MTM VAR=RATIO STRATA=STG/N3,B1/DELTA,B2/THETA CH=${cen}
b=1.3862 m=1.2243 sd=0.4581
That is, this feature is a ratio between two band powers (delta and
theta) from N3 sleep, averaged across all central channels listed in
the variable ${cen} (note, this variable does not need to be
labelled ${cen}, it could be any valid Luna variable name,
e.g. ${s}). The label name (delta_theta_mean_C_N3) is aribitrary,
used only to identify the feature, i.e. it could equally be ftr01
etc. (It should not contain spaces or an equals sign, however.) Here,
the feature is described across multiple lines, although it is also
permissable to use a single line. The label must come first; all other attributes,
that will be in the form of key=value pairs, can be in any order.
Using the cache
The cache is a mechanism whereby one Luna command can pass information to another Luna command during the processing of a single recording - i.e. a temporary store specific to an individual.
When used with PREDICT, there are three primary points
at which the cache is invoked:
-
via the
CACHE recordstatement, to tell Luna which metrics to track -
when a tracked command is run, any tracked values will be cached
-
when
PREDICTruns, based on the model specification file details, values will be pulled from the cache and used to build the vector of predictors for that individual
To take the specific case above, the first component might be:
CACHE cache=c1 record=MTM,RATIO,B1,B2,CH,STG
The record argument expects values in the form/order: command,
variable, one or more strata. This names a cache c1 (i.e. the
same cache will be passed to PREDICT) and instructs Luna to cache
any RATIO values emitted by the MTM command that have the
associated strata of B1, B2, CH and (in this particular case)
STG. Here, RATIO is the ratio of two band powers (B1 / B2 ),
which will be defined for a given channel CH. By default, any
RATIO variable will always have at least these three associated
strata, B1, B2 and CH: e.g. B1=DELTA, B2=ALPHA and
CH=C3_M2. Here, we additionally specify a stratifying factor of
STG, corresponding to sleep stage. This is a user-defined strata
specified via the TAG command, used to track
N2 versus N3 metrics calculated in the same script. The tag makes N2
and N3 metrics distinct, otherwise new calls to MTM would overwrite
the output associated with previous ones in the same run.
Given the above CACHE command, Luna will cache those values from any
outputs that match all these conditions (i.e. for that command,
variables and strata combination):
MASK ifnot=N3
RE preserve-cache
TAG STG/N3
MTM sig=C3,C4 ratio1 ratio=DELTA/THETA,DELTA/ALPHA
After running the MTM command above, Luna will cache the four
following strata, each defined for four factor/level pairs, for the
RATIO variable (that is, a single number representing the power
ratio for that channel and pair of bands for N3 sleep):
CH/C3B1/DELTAB2/THETASTG/N3CH/C4B1/DELTAB2/THETASTG/N3CH/C3B1/DELTAB2/ALPHASTG/N3CH/C4B1/DELTAB2/ALPHASTG/N3
The final step involving the cache is when PREDICT retrieves
cached values for that recording, given a set of feature definitions, e.g.:
delta_theta_mean_C_N3
CMD=MTM VAR=RATIO STRATA=STG/N3,B1/DELTA,B2/THETA CH=${cen}
b=1.3862 m=1.2243 sd=0.4581
Luna will pull the first two of the four values cached above, as both
match the level values for the factors B1, B2 and STG. The
channel specification (CH) is handled separately from the other
strata (which are specified by the STRATA keyword). Assuming the
${cen} was previously defined to be cen=C3,C4, this will retrieve
estimates of N3 delta/theta power ratio from both channels and then
take the average. The CH keyword can handle one or multiple
channels, but will always emit a single (averaged) value.
Restructing/freezing with caches
In the above example, note that the RE
command had the special option preserve-cache. By default,
RE and THAW would
otherwise wipe the cache. When using a cache, this is typically not
what one wants, i.e. if we wish to retain the cached values until a
subsequent PREDICT command.
Consider the following example (given here not as a full working
example, but just a skeletal script): if we wished to use sigma band
power from both N2 and N3 (from the Welch PSD command, which emits a
variable also called PSD with strata defined by band B and channel CH alongside
a further STG stratum that aligns with the TAG commands below):
CACHE cache=c1 record=PSD,PSD,B,CH,STG
% pre-process whole signal
FILTER sig=CZ bandpass=0.3,35 tw=1 ripple=0.01
FREEZE F1
% get N2 metrics
TAG STG/N2
MASK ifnot=N2
RE preserve-cache
PSD sig=CZ
THAW tag=F1 preserve-cache
% get N3 metrics
TAG STG/N3
MASK ifnot=N2
RE preserve-cache
PSD sig=CZ
% clear STG, else PREDICT output would have a STG/N3 stratum
TAG STG/.
% predict given both N2 and N3 metrics
PREDICT cache=c1 model=model1.txt
In the above, the cached metrics from N2 would be lost if we thawed
the previous F1 freeze (i.e. as that freeze did not contain the cached
values). That is, the preserve-cache option decouples the cache
from the typical snapshot mechanism for returning to a former state.
A similar logic applies with the restructure command. The simple rule
is: if building a script that uses PREDICT and a cache,
add preserve-cache to THAW and RE.
Using variables
In the above example, channel CH is set to a Luna variable (here
${cen}), which allows more flexibility, i.e. one does not need to
edit the model specification file if running on a dataset with a
different label.
In addition, to include features in a predictive model that are not
derived from Luna commands per se (and will therefore not be in the cache),
you can use the VALUE keyword when defining a model: e.g. to include
a covariate for male versus female sex:
male_sex
VALUE=${male} b=-0.22 m=0.48 sd=0.5
This assumes that a variable called ${male} will have been defined
for that individual (even if it is defined as missing for that
individual). Such a variable is specified in the same way as any other
Luna variables, e.g. on the command line
luna file1.edf male=1 -o out.db < predict.txt
luna s.lst vars=covar.txt -o out.db < predict.txt
covar.txt is a tab-delimited file, with ID as the first column, and a column labelled male (note: case-sensitive)
as one of the other columns:
ID age male site
id001 12 1 A
id002 16 0 A
id003 9 . B
id004 6 1 B
...
If you specify a VALUE term for a particular feature, you should not
also specify CMD, VAR or STRATA terms as well, as those relate
to searching the cache.
Special variables
As well as defining the core terms, the model specification file also specifies a few special variables, in the format
variable <- value
intercept <- 42
The primary special variables are
| Variable | Description |
|---|---|
observed |
If known, the observed value (e.g. chronological age) to be used in output, and bias-adjustment |
intercept |
The model intercept |
data |
The filename of the feature matrix data file (for kNN imputation) - can also be given as data as a option to PREDICT |
knn |
The number of nearest neighbours to consider when running kNN imputation |
minf |
The minimum number of non-missing features required to run the model |
softplus |
Apply the softplus function to the output (0/1=N/Y) |
log1p |
Apply log1p() scaling to all inputs (0/1=N/Y) |
There are also some special labels expected that can be used to describe the model (currently not directly used by Luna, but adding to the model file can help to document the model for users):
| Variable | Description |
|---|---|
title |
A title for the model |
reference |
A PMID or similar reference for the model/data |
outcome |
The type/unit of the predicted outcome, e.g. Age (years) |
type |
Type of model (linear/logistic) - currently only linear models supported |
training |
Brief note about the training data or procedure |
Bias-adjustment models
Some models (including Sun et al. mentioned above) require a bias-adjustment term (e.g. as described here).
| Variable | Description |
|---|---|
bias_correction_slope |
Coefficient for bias adjustment model (b) |
bias_correction_intercept |
Intercept (c) |
bias_correction_term |
Observed/known value (e.g. chronological age), same as ${observed} (a) |
If these terms are found in a model specification file, then a
bias-adjusted version of the prediction (Y1) will be output as well
as the raw estimate (Y).
This particular form of bias adjustment uses the observed value of the output variable, as is appropriate in the context of biological/brain age prediction models, i.e. where chronological age will be known, and it is the difference between predicted and chronological age that is the outcome of primary interest.
Given b, c and a as defined above, and an initial prediction y, the bias-adjusted value y1 is simply defined as
Y1 = Y - ( B * A + C )
If PREDICT is given the above terms in the model file (along with
the observed value A), it will automaticall calculate and output
Y1 as well as Y.
Reference data
If a reference dataset is included with the data option (to support kNN imputation) it must have the following format:
-
each row is one individual/observation, each column is one feature; all features are whitespace-delimited
-
first row starts with
#followed by number of rows (observations) and columns (features), e.g.# 2936 13 -
second row also starts
#and then lists all feature labels (that must match the terms in the model file exactly) -
subsequent rows contain the numeric values, expected to be standardized (i.e. each column has mean of 0, SD of 1)
See this file for an example.
PREDICT
Make a prediction based on a specified model and cached Luna metrics
See the overview above for a high-level description of
this command, and how it fits into a broader paradigm of using Luna
to support model-based prediction. PREDICT is typically not used alone, but
rather needs to be paired with a) a model specification
and b) a Luna script with upstream commands to compute and cache
the features (predictors) used in the model.
Internally, the steps are:
-
PREDICTreads the model specification file and swaps in any variables (similarly to how Luna parses command scripts); this is done separately for each individual, i.e. meaning that model files can contain variables that vary between different individuals -
it then attempts to find values for the specified model features; typically these will be from the cache, but if a variable exists with (exactly) the same name (i.e from a vars file) then it will be used; otherwise, the default is for
PREDICTto search the cache -
next, it checks there are enough non-missing features, as specified by
REQorminfin the model file -
it then standardizes all features based on population mean/SD values (which are always included in a model file)
-
PREDICTuses a simple k-nearest neighbour (kNN) approach to impute missing values, based onknn=10neighbours by default; kNN imputation is only performed if a referencedatafile has been attached -
if a reference dataset is available,
PREDICTalso uses it to identify outlier features, by dropping each non-missing feature in turn, re-imputing it, and then calculating the difference (observed - imputed) in SD units. If the distance exceeds theththreshold set, the observed values are taken to be outliers, and replaced with their imputed values -
using standardized features,
PREDICTthen makes a prediction based on the implied linear model; if the model file specifies it, a subsequent bias-adjustment procedure is applied (see above)
Cacheless mode
If features have been pre-computed and are all available in a simple text file, PREDICT can run
quickly in cacheless mode, i.e. directly taking feature values as variables rather than running
Luna commands on the raw data. In this way, you could use PREDICT as a standalone command (on
the assumption that features.txt has columns defined to match each defined term in model.txt):
luna s.lst vars=features.txt -o out.db -s PREDICT model=model.txt
Parameters
Primary parameters
| Option | Example | Description |
|---|---|---|
model |
models/m1.txt |
Model/feature specification filename |
cache |
p1 |
Specify a prior cache name |
data |
models/m1.data |
Data set (to support kNN imputation) |
th |
4 | Absolute Z-value distance to trigger re-imputation |
Secondary parameters
| Option | Example | Description |
|---|---|---|
drop |
ftr2,ftr3 |
Drop one or more features from the model |
dump-model |
Dump the model to standard output |
Outputs
Individual-level output (strata: none):
| Variable | Description |
|---|---|
NF |
Number of features |
NF_OBS |
Number of features observed (non-missing) |
OKAY |
Flag for whether sufficient non-missing features were observed (0/1=N/Y) |
Y |
Default prediction |
Y1 |
Optional, bias-adjusted prediction |
YOBS |
Observed value for outcome, if known |
Feature-level output (strata: FTR)
| Variable | Description |
|---|---|
X |
Raw value of this feature |
Z |
Normalized feature value |
D |
kNN-derived distance for any non-missing feature |
IMP |
Flag for whether this feature was imputed, i.e. if missing (0/1=N/Y) |
REIMP |
Flag for whether this feature was "re-imputed"(*), i.e. if an outlier (0/1=N/Y) |
B |
Feature coefficient (from the model, fixed for all individuals) |
M |
Feature mean (from the model, fixed for all individuals) |
SD |
Feature standard deviation (from the model, fixed for all individuals) |
(*) Calling this 're-imputed' doesn't really make much sense, but the output is stuck with this nomenclature for now.
Example
As a full working example, we will consider the model described in Sun et al (2019). This model contains 13 features based on the NREM sleep EEG. It was trained on 2,532 individuals to predict an individual's age.
-
The model specification file is available here.
-
The Luna script used to extract the features and run the
PREDICTcommand is available here. -
The above repository also contains the datafile for kNN imputation here.
For simplicity, imagine these files are called m-features.txt,
m-luna.txt and m-data.txt respectively, available (relative to the
current working folder) in the folder models/.
The Luna script a) sets up the cache, b) does some pre-processing, c)
extracts metrics for N1, then N2, then N3 sleep, using the freeze/thaw
mechanism to swap between stages, and then d) runs PREDICT to make a predicton.
If p1.lst is a sample list pointing to the EDF and staging
annotations for one 69 year-old individual, then we could run:
luna p1.lst age=69 cen=C3,C4 th=3 mpath=models/ -o out.db < m-luna.txt
The Luna script expects the variables ${th} and ${mpath} to be
defined, as these are passed as parameters to the PREDICT command,
as well as ${cen}, to indicate which (central mastoid-referenced
EEG) channels to derive metrics from. The model specification file
further expects the variables ${age} and ${cen} to be defined. In
practice, ${age} (which obviously varies between individuals) would
be specified via a vars file.
Re-referencing on-the-fly
Note that in this particular case, the channels were not contra-lateral mastoid referenced, whereas the above m-luna.txt
assumes they are (inspect the script to see). Rather than edit the script, or use two versions, one can splice in additional
commands to Luna, by using standard command-line tools: i.e. what we actually ran was (the other arguments replaced with ... here):
echo "REFERENCE sig=C3 ref=A2 & REFERENCE sig=C4 ref=A1" | cat - m-luna.txt | luna p1.lst ...
luna p1.lst ... < m-luna.txt
m-luna.txt to include the extra REFERENCE commands. The above is equivalent to:
cat m-luna.txt | luna p1.lst ...
cat - m-luna.txt to concatenate the standard input (here from a prior echo) with the Luna script, and
then all of that gets piped into Luna, i.e. in the form:
echo "extra first commands go here" | cat - script.txt | luna p1.lst ...
This script contains multiple commands and generates a lot of console
output. It is always worth reviewing in test cases that the script it
performing as expected. The final PREDICT command gives the
following messages to the console:
CMD #37: PREDICT
options: cache=p1 data=models/m-data.txt model=models/m-features.txt sig=* th=3
read 13 terms and 8 special variables from models/m-features.txt
creating 2936 x 13 reference feature matrix from models/m-data.txt
applying softplus scaling to predicted values
predicted value (Y) = 59.8799
bias-corrected predicted value (Y1) = 70.613
observed value (YOBS) = 69
That is, this individual had an observed age of 69 years, and a (bias-adjusted) estimated age (based on the NREM EEG) of 70.6 years.
The primary outputs are available in out.db:
destrat out.db +PREDICT
ID NF NF_OBS OKAY Y Y1 YOBS
id01 13 13 1 59.879 70.613 69
where NF is the number of feautres used in the model. Depending on
the model used, either Y or Y1 should be considered as the primary
output.
We can view the individual features:
destrat out.db +PREDICT -r FTR
ID FTR B M SD X D Z IMP REIMP
id01 COUPL_OVERLAP_C -0.804 366.3 191.7 270.0 -0.178 -0.502 0 0
id01 DENS_C -1.665 4.513 1.911 3.486 -0.857 -0.537 0 0
id01 alpha_bandpower_kurtosis_C_N2 -3.184 7.331 2.598 6.263 -0.108 -0.411 0 0
id01 alpha_bandpower_mean_C_N1 2.291 0.068 0.047 0.046 -1.395 -0.465 0 0
id01 delta_alpha_mean_C_N3 -1.348 1.344 0.548 0.808 -0.266 -0.975 0 0
id01 delta_bandpower_kurtosis_C_N2 -1.868 17.01 4.071 10.47 -0.496 -1.607 0 0
id01 delta_bandpower_mean_C_N3 -2.620 1.445 0.618 0.857 -0.279 -0.949 0 0
id01 delta_theta_mean_C_N3 1.386 1.224 0.458 0.744 -0.310 -1.049 0 0
id01 kurtosis_N2_C -0.052 2.851 1.349 1.362 -0.047 -1.103 0 0
id01 kurtosis_N3_C -1.247 1.086 0.576 0.335 -0.368 -1.302 0 0
id01 sigma_bandpower_kurtosis_C_N2 1.247 15.19 4.749 18.11 0.741 0.615 0 0
id01 theta_bandpower_kurtosis_C_N2 -3.744 7.461 2.557 6.222 0.440 -0.484 0 0
id01 theta_bandpower_kurtosis_C_N3 0.157 5.364 2.045 2.301 -0.962 -1.496 0 0
-
the first three give the coefficient, mean and population SD and will be identical for all individuals (i.e. these are directly from the model file)
-
Xis the observed value of the feature (in this case, all derived from cached Luna commands) -
Dis the distance in SD units for the expected value of the feature based on kNN imputation based on all non-missing features but excluding this one -
Zis the normalized (potentially imputed) final version used in the prediction equation (i.e. multipled byB) -
IMPandREIMPindicate whether the finalZvalue was imputed, either because it was missing (IMP) or an outlier based onD(REIMP)
If particular features appear to be consistently noisy or biased, they
can be dropped from the model by adding the option drop=F1,F2 where
F1 and F2 are two features, for example.
Building feature matrices to re-run models
If a script takes a long time to compute features, or if you have a large sample, and you wish to re-run models (e.g. changing parameter settings) it can be advantageous to extract all features from the output to make a single text input file.
We first pull the values X (i.e. the raw features used by the model, but after averaging across
channels) as follows:
destrat out.db +PREDICT -c FTR -v X > ftr.txt
ID X.FTR_COUPL_OVERLAP_C X.FTR_DENS_C ...
id01 270 3.48598130841121 ...
ID field) but the labels (following default destrat output practices have the variable/factor name X.FTR_ at the front of each column label. We can strip these, either manually, as using something as follows:
sed 's/X\.FTR_//g' < ftr.txt > ftr2.txt
ID COUPL_OVERLAP_C DENS_C ...
id01 270 3.48598130841121 ...
X.FTR_ etc, but this works for now.
We can then re-run the single PREDICT step as follows:
luna p1.lst vars=ftr2.txt age=69 cen=C3,C4 -o out2.db -s PREDICT model=models/m-features.txt data=models/m-data.txt th=3
bias-corrected predicted value (Y1) = 70.613
observed value (YOBS) = 69
Note that in practice, age (and potentially other information such as the central channel labels to use) could instead be
kept in a file (with multiple individuals on different rows), e.g. covar.txt
ID age male cen
id01 69 1 C3,C4
id02 73 0 CZ
id03 56 1 C3,C4
...
luna p1.lst vars=ftr2.txt,covar.txt -o out2.db -s PREDICT model=models/m-features.txt data=models/m-data.txt th=3
Now we can easily change parameters, e.g. varying th or dropping particular terms. For example, here we might drop both spindle-related
metrics: running the same command as above but adding to PREDICT
drop=DENS_C,COUPL_OVERLAP_C
bias-corrected predicted value (Y1) = 68.6742
observed value (YOBS) = 69
destrat out2.db +PREDICT -r FTR -v IMP X Z
ID FTR IMP X Z
id01 COUPL_OVERLAP_C 1 NA -0.015
id01 DENS_C 1 NA 0.391
id01 alpha_bandpower_kurtosis_C_N2 0 6.263 -0.411
id01 alpha_bandpower_mean_C_N1 0 0.046 -0.465
id01 delta_alpha_mean_C_N3 0 0.809 -0.976
id01 delta_bandpower_kurtosis_C_N2 0 10.47 -1.607
id01 delta_bandpower_mean_C_N3 0 0.858 -0.950
id01 delta_theta_mean_C_N3 0 0.744 -1.049
id01 kurtosis_N2_C 0 1.363 -1.103
id01 kurtosis_N3_C 0 0.335 -1.302
id01 sigma_bandpower_kurtosis_C_N2 0 18.11 0.615
id01 theta_bandpower_kurtosis_C_N2 0 6.222 -0.485
id01 theta_bandpower_kurtosis_C_N3 0 2.302 -1.497
The NA for X in the top two rows indicated they were dropped from
the original model as observed variables, i.e. set to missing then
imputed. As expected these values deviate from the observed values
somewhat : -0.015 and 0.391 instead of -0.502 and -0.537 and have
resulted in a slightly lower estimated age. In a larger sample, one could use the
above framework to perform sensitivity analyses, etc, e.g. to see if some terms increase or
decrease noise in predictions significantly.
Viewing the original channel-level features
Finally, just to connect the features (internally cached metrics
passed to PREDICT) with the "standard" Luna outputs, we can look at
the rest of the out.db file, which will contain the same values that
were cached. The one difference is that PREDICT internally averaged
over multiple channels, and so we'll need to do that here to check
that things line up.
To take a few examples: first spindle density, which has the label DENS_C in the model:
destrat out.db +PREDICT -r FTR/DENS_C -v X
ID FTR X
id01 DENS_C 3.48598
From the main SPINDLES output, we know it was stratified by F and
CH (as always for spindle density) but also STG (because this was
added in the script via TAG STG/N2 before the spindles command):
destrat out.db +SPINDLES -r F CH STG -v DENS
ID CH F STG DENS
id01 C3 13.5 N2 3.65421
id01 C4 13.5 N2 3.31776
As expected, the mean of these two values equals the value of X from
PREDICT, i.e. (3.65421 + 3.31776)/2 = 3.48598.
To consider a second example: the delta/alpha N3 power ratio: delta_alpha_mean_C_N3. From the model file,
we can see the definition:
delta_alpha_mean_C_N3
CMD=MTM VAR=RATIO STRATA=STG/N3,B1/DELTA,B2/ALPHA CH=${cen}
RATIO variable of the MTM command, and based on a stratum defined by the two power bands, a stage and channels:
destrat out.db +MTM -r B1/DELTA B2/ALPHA STG/N3 CH -v RATIO
ID B1 B2 CH STG RATIO
id01 DELTA ALPHA C3 N3 0.78152
id01 DELTA ALPHA C4 N3 0.83614
X to be the mean of these, namely
(0.78152+0.83614)/2 = 0.80883. From PREDICT itself, the averaged
value was:
destrat out.db +PREDICT -r FTR/delta_alpha_mean_C_N3 -v X
ID FTR X
id01 delta_alpha_mean_C_N3 0.8088296
Models
Currently, in its initial release, only a single model is supported - more will be added soon.
| Label | Model | Link |
|---|---|---|
| SUN2019 | "Brain-age" prediction for adults | repo |
SUN2019
Sun et al (2019) uses 13 NREM sleep EEG features to provide a robust estimate of "brain age". The difference between predicted and observed age is labelled the brain age index. The model was trained on over 2,500 adults aged 18 to 80.