Docker
The Docker distribution of Luna is designed to allow users to test both the lunaC and lunaR, that is, the C/C++ command-line and the R package versions of Luna. Although beyond the scope of this documentation, Docker containers can also facilitate using Luna in a cloud computing environment, such as Amazon Web Services EC2.
What is Docker?
Docker is a popular tool, increasingly used in cloud-computing, that is used to deploy and run applications on diverse host platforms (i.e. including your computer). These Getting Started pages will help to orient you, although most users of Luna need not read past the first page. From the perspective of a user of Luna, all you'll need to do is download the free Desktop version of Docker CE (Community Edition) and then use one or two commands to launch the containerized version of Luna (which includes a full Linux environment comprising lunaC, lunaR, the tutorial data as well as R or RStudio. For those interested in learning more about Docker, this tutorial is a good place to start.
Available images
We currently support three main Docker images:
-
remnrem/lunalite: just the command-line executables (luna,destratandbehead) in a lightweight container -
remnrem/luna: Luna, lunaR and RStudio along with the tutorial data -
remnrem/lunapi: Luna, lunaR as well as the Python-based lunapi wrapper for Luna, including a JupyterLab notebook environment, as described here
Getting Docker
Docker provides a quick way to run Luna on any system capable of installing Docker. Docker containers are similar in principle to virtual machines that can run on any platform Docker is installed on.
Desktop versions of Docker can also be installed (for free) on both Mac and Windows by following either one of those links. Note that the latter requires Windows 10 Professional or Enterprise 64-bit. Follow the extensive documentation on the Docker website to get up-and-running with Docker.
Docker is available for most Linux machines: for example, here
using apt-get on an Amazon Linux EC2 instance:
sudo apt-get install docker
Hint
Versions of Docker for older Windows or Mac machines are also available, although we have not tried these and cannot give advice.
Pulling luna
Once you have Docker running on your system (the start up can take a minute or two after installation), the following command (typed in a terminal/command prompt) will download an image (from Docker Hub) and create a containerized version of Luna:
docker run --rm -it remnrem/luna /bin/bash
(Alternatively, swap in remnrem/lunalite for the lightweight version.)
The remnrem/luna Docker image contains luna pre-installed within a
Linux environment. It also includes R with lunaR
pre-installed: both base R and RStudio, as
described below.
You should see something like the following:
Unable to find image 'remnrem/luna:latest' locally
latest: Pulling from remnrem/luna
6cf436f81810: Pull complete
987088a85b96: Pull complete
b4624b3efe06: Pull complete
d42beb8ded59: Pull complete
166613f8d770: Pull complete
d22664acc861: Pull complete
dc715f416e3a: Pull complete
4cb433a57b07: Pull complete
02ec624d4949: Pull complete
Digest: sha256:95da9c1a6e2cc7d88123c3bc53776768605fc4e94ae6d058e078b437eb2efd1b
Status: Downloaded newer image for remnrem/luna:latest
root@82bcf5c52204:/data#
The last line of the output above is in fact a prompt running
inside a newly created container. (In Docker terminology, a
container is a particular instantiation of an image.) This container provides a
Linux command line (based on Ubuntu Linux), which has Luna pre-installed. That is, if
you type luna at the command line, you should see:
root@82bcf5c52204:/data# luna
===================================================================
+++ luna | v0.9, 12-Feb-2019 | starting 11-Mar-2019 19:46:07 +++
===================================================================
usage: luna [sample-list|EDF] [n1] [n2] [@parameter-file] [sig=s1,s2] [v1=val1] < command-file
This Luna image also has the tutorial datasets under
the /tutorial/ folder:
root@82bcf5c52204:/data# ls /tutorial/
edfs s.lst
The s.lst file is the sample-list
that describes a project consisting of the three tutorial individuals:
root@82bcf5c52204:/data# cat /tutorial/s.lst
nsrr01 edfs/learn-nsrr01.edf edfs/learn-nsrr01-profusion.xml
nsrr02 edfs/learn-nsrr02.edf edfs/learn-nsrr02-profusion.xml
nsrr03 edfs/learn-nsrr03.edf edfs/learn-nsrr03-profusion.xml
You can run Luna and perform the tutorial actions within this environment: (note: as the
s.lst file uses relative paths you need to add the path
on the command line):
root@82bcf5c52204:/data# luna /tutorial/s.lst 1 path=/tutorial -s DESC
===================================================================
+++ luna | v0.9, 12-Feb-2019 | starting 11-Mar-2019 20:00:14 +++
===================================================================
input(s): /tutorial/s.lst
output : .
commands: c1 DESC
path : /tutorial/
___________________________________________________________________
Processing: nsrr01 [ #1 ]
total duration 11:22:00, with last time-point at 11:22:00
40920 records, each of 1 second(s)
signals: 14 (of 14) selected in a standard EDF file:
SaO2 | PR | EEG(sec) | ECG | EMG | EOG(L) | EOG(R) | EEG
AIRFLOW | THOR RES | ABDO RES | POSITION | LIGHT | OX STAT
annotations:
[NREM1] 109 instance(s) (from /tutorial/edfs/learn-nsrr01-profusion.xml)
[NREM2] 523 instance(s) (from /tutorial/edfs/learn-nsrr01-profusion.xml)
[NREM3] 16 instance(s) (from /tutorial/edfs/learn-nsrr01-profusion.xml)
[NREM4] 1 instance(s) (from /tutorial/edfs/learn-nsrr01-profusion.xml)
[REM] 238 instance(s) (from /tutorial/edfs/learn-nsrr01-profusion.xml)
[apnea_obstructive] 37 instance(s) (from /tutorial/edfs/learn-nsrr01-profusion.xml)
[arousal] 194 instance(s) (from /tutorial/edfs/learn-nsrr01-profusion.xml)
[artifact_SpO2] 59 instance(s) (from /tutorial/edfs/learn-nsrr01-profusion.xml)
[desat] 254 instance(s) (from /tutorial/edfs/learn-nsrr01-profusion.xml)
[hypopnea] 361 instance(s) (from /tutorial/edfs/learn-nsrr01-profusion.xml)
[wake] 477 instance(s) (from /tutorial/edfs/learn-nsrr01-profusion.xml)
..................................................................
CMD #1: DESC
EDF filename : /tutorial/edfs/learn-nsrr01.edf
ID : nsrr01
Duration : 11:22:00
# signals : 14
Signals : SaO2 PR EEG(sec) ECG EMG EOG(L) EOG(R) EEG AIRFLOW THOR RES ABDO RES POSITION LIGHT OX STAT
___________________________________________________________________
...processed 1 EDFs, done.
...processed 1 command set(s), all of which passed
-------------------------------------------------------------------
+++ luna | finishing 11-Mar-2019 20:00:14 +++
===================================================================
This container also provides the R package with the lunaR library pre-installed. To start R:
root@82bcf5c52204:/data# R
To attach the lunaR library:
> library(luna)
** lunaR v0.2 1-Mar-2019
** luna v0.9 12-Feb-2019
To attach the tutorial project:
> sl <- lsl( "/tutorial/s.lst" , path = "/tutorial/" )
3 observations in /tutorial/s.lst
To attach, for example, the second individual nsrr02:
> lattach( sl , 2 )
nsrr02 : 14 signals, 10 annotations, 09:57:30 duration
Container contents
-
mount points:
/data/,/data1/and/data2/allow different folders on the host machine to be mapped to these folders within the container -
text editors:
emacs(powerful but complicated to use if you are not familiar with it) andnano(simple) are pre-installed -
build directory:
/build/, this folder container the source code used to build lunaC and lunaR, and can be ignored by most users
Luna and RStudio
As well as the basic container remnrem/luna, we also have generated
an image that bundles RStudio with a pre-installed version of lunaC
and lunaR. To obtain this:
docker run -e PASSWORD=abc123 -p 8787:8787 remnrem/luna
If this command is successful, then after it downloads and installs, you will be able
to go to you web browser and visit the URL: localhost:8787
Log in with username name rstudio and the password you specified above (i.e. abc123). You will
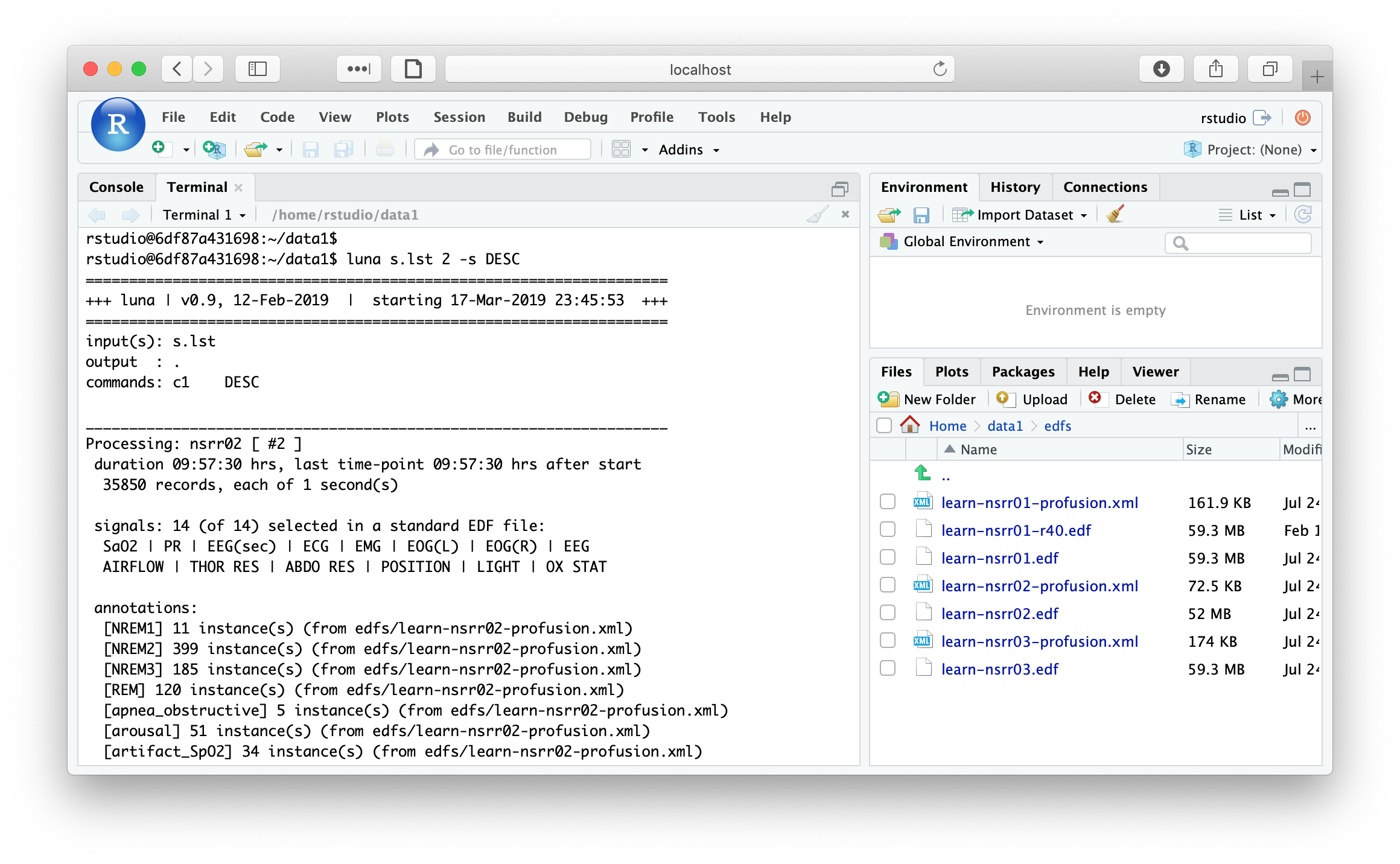
then have an interactive environment, for which both the command prompt (for lunaC) and R (for lunaR)
are available, along with the tutorial data. For example, we can run lunaC in the terminal:
Likewise, we can use lunaR to query the tutorial data:
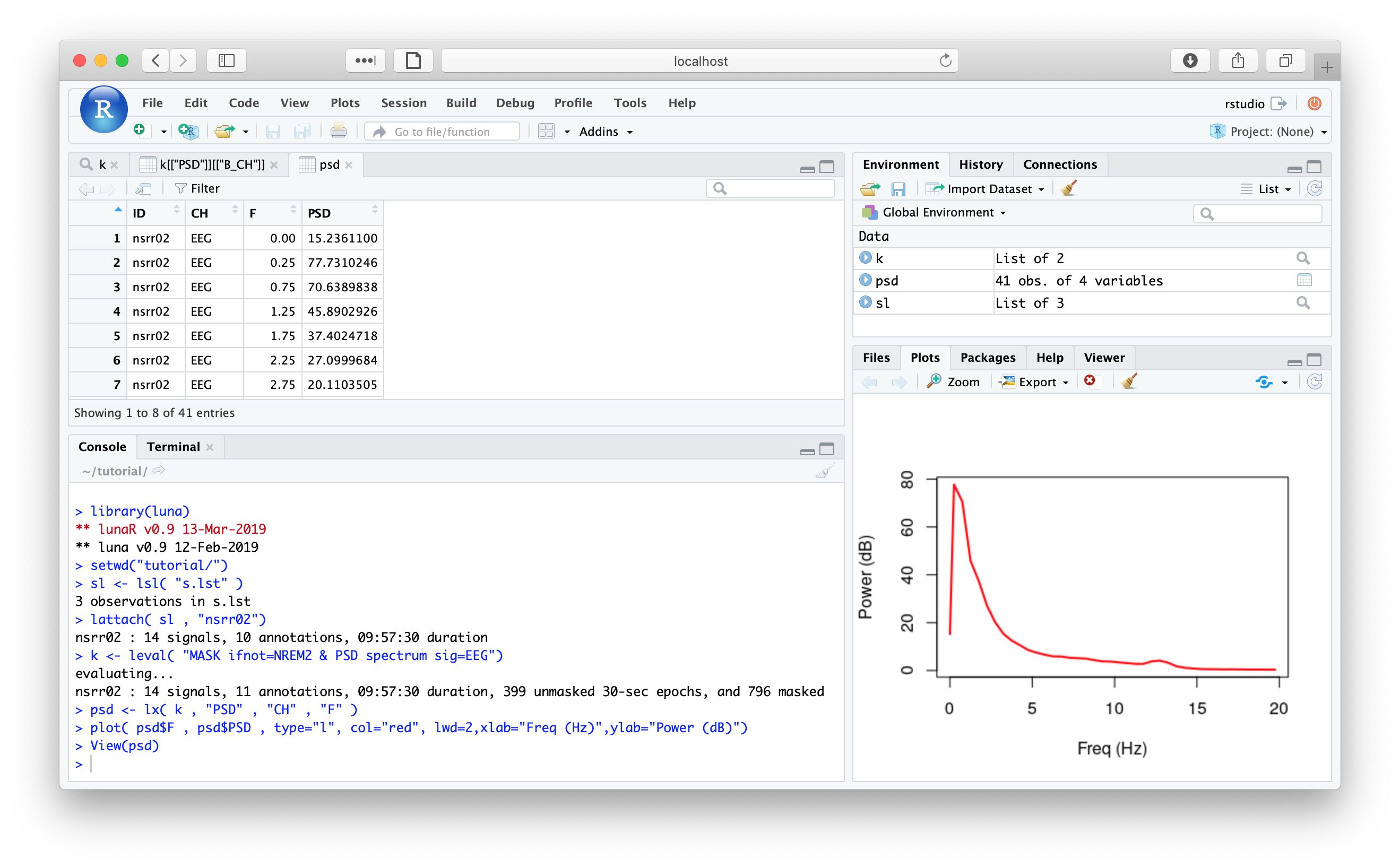
The commands for mapping local folders to the Docker container (i.e. so that your own data are visible whilst in the container, and so that your output will be saved back on the host machine) are the same as for the standard Luna image.
luna.docker
For Linux and Mac OS machines with the
bash shell
installed, we provide a convenience wrapper script to help starting
new Luna containers (i.e. instead of running docker run etc). See
Docker documentation for full
details on using Docker.
Download the script luna.docker from
here, save it in
your path (e.g. see echo $PATH) and set permissions to be executable, e.g.:
chmod +x /usr/local/bin/luna.docker
Note that, depending where you place it and your operating system, you
may need to prefix this (and subsequent) docker command with sudo.
Typing luna.docker will then show the usage of this script:
new instance : luna.docker /path/data/ /path/data1/ /path/data2/
new RStudio instance : luna.docker -rs password /path/data/ /path/data1/ /path/data2/
list instances : luna.docker -l
attach to running instance : luna.docker -a {container-ID}
delete instance : luna.docker -d [container-ID|all]
pull latest luna image : luna.docker --pull
To start a new instance, specify between one and three paths, that
will be mounted to /data/, /data1/ and /data2/ respectively (or,
with RStudio, in /home/rstudio/data/, etc).
Hint
Although the paths should generally be full/absolute paths
(i.e. starting /), for the first argument only, the period (.)
can be used to mount /data/ to the current working directory.
For example, if on the host system we are in the folder test:
$ pwd
/Users/joe/test
$ ls
a.txt b.txt c.txt
luna.docker .
mounting /Users/joe/test to /data/
/data/ folder; therefore, running ls will show the contents of /Users/joe/test/ but from inside the container:
root@ab8e676352ec:/data# pwd
/data
root@ab8e676352ec:/data# ls
a.txt b.txt c.txt
Any new files you create in /data/ from within the container will
also be visible in /Users/joe/test/ after leaving the container
(with Control-D) or typing exit. For example, from within the container
root@ab8e676352ec:/data# echo "Hello World!" > d.txt
$ ls
a.txt b.txt c.txt d.txt
cat d.txt
Hello World!
In this way, you can map inputs (i.e. EDFs and annotation files) to
outputs (e.g. destrat databases or extracted plain-text files) between
your host system and the container. Being able to specify multiple
mounted volumes (on /data/, /data1/ and /data2/) is convenient
if you have different EDFs in different locations, or if you want
output to be written to a different folder than contains the input
EDFs, etc.
Other options
You can initiate multiple containers at the same time. Under standard conditions, different containers are completely isolated: i.e. new files generated in one container will not be present in other containers, etc.
Running with the -l flag will list the currently active containers:
luna.docker -l
CONTAINER ID IMAGE COMMAND CREATED NAMES
e07c4e1adca2 remnrem/luna "/bin/bash" 3 seconds ago serene_bartik
To attach a new shell on an existing running container, use the -a
option, along with the container ID or NAME. In the above case,
the ID is e07c4e1adca2 and the NAME is serene_bartik (Docker
automatically generates cute names.) For example, this will open a new
terminal command prompt for the running container:
luna.docker -a serene_bartik
This can be useful if you have a long-running process in one container and want to check its output, or if you want to have a text-editor open in one window and cut-and-paste to run those commands in another window, for example.
To delete an active container, use the -d flag, followed by either
the ID or NAME of that container. For example:
luna.docker -d e07c4e1adca2
removing e07c4e1adca2...
e07c4e1adca2
-l option now shows that there are no running containers:
luna.docker -l
CONTAINER ID IMAGE COMMAND CREATED NAMES
As noted, this doesn't even begin to scratch the surface of using
Docker containers, a full treatment of which is well beyond the scope
of this documentation. For example, you are able to control the
memory and CPU cores available for each Docker container (simply via
the Preferences... menu of the Desktop versions of Docker). This
will likely be important if running larger analysis jobs with Luna.
We provide a containerized version of Luna as an experimental feature, but have not yet used it extensively ourselves: please let us know if you encounter limitations or if you think there are better ways to set things up...
Windows machines
It is possible to run Docker on recent Windows machines, although you
may need to consult the Docker website for exact information. As the
luna.docker script above will not work for Windows/DOS, you'll need
to type the exact Docker commands, including mapping any volumes or
ports between the host and the container, as detailed below.
LunaR
At the Command Prompt, to open a new container and map the D:\luna
folder on the host to /data/ in the container, and C:\mydata\work to /data1 type:
docker run --rm -it -v D:/luna:/data -v C:/mydata/work:/data1 remnrem/luna /bin/bash
By default, the container has folders /data, /data1 and /data2
to allow three volumes to be mounted, i.e. folders on the host machine
to be shared with the Luna container.
LunaRStudio
At the Command Prompt, to open a new container and map the D:\luna
folder on the host to /data/ in the container, type:
docker run --rm -e PASSWORD=abc123 -p 8787:8787 -v D:/luna:/data remnrem/luna
and then open your web-browser at http://localhost:8787 and enter
username rstudio and password abc123.
Listing/killing/attaching to containers
To list all containers:
docker ps
To clean up everything:
docker system prune
To kill a particular container, e.g. with name kind_herschel
docker container rm -f kind_herschel
To open another terminal/connection to an existing container, e.g. called kind_herschel
docker exec -i -t kind_herschel bash -l
Hints
-
Docker takes a while (minutes) to start running, so be sure that it is running before you try to launch Luna
-
anecdotally, on Windows I've had issues getting any Docker command to run that were solved by simply restarting Docker (i.e. look to the Docker small icon in your task bar)
-
if you have problems, try Docker's hello-world example/tutorial
-
expect to run into issues if your institution/machine has particular firewalls or other types of network security features, of if you are trying to to mount folders on access-protected shared volumes, etc. These types of issues are completely beyond the scope of Luna and this documentation, and things you'll need to work with your IT team to fix
-
the whole point of Luna Docker is that it is meant to provide a quick, out-of-the-box way to run/test Luna. If you are having trouble trying using a binary version, or a different operating system.
-
in general, you'll likely have a better Luna experience if you use a Unix-like operating system, i.e. macOS or Linux. Of course, use whatever works best for you, but if you're working with large amounts of data there are numerous reasons to consider alternatives to Windows that may work better...
remnrem/lunalite
To obtain the lightweight version:
docker pull remnrem/lunalite
To run Luna from the host:
docker run --rm -it remnrem/lunalite luna -v
luna-base version v1.1.0 (release date 11-Oct-2024)
luna-base build date/time Oct 13 2024 22:06:18
Eigen library v3.4.0
sqlite v3.41.2
To run interactively:
docker run --rm -it remnrem/lunalite
/data # is part of the shell prompt here, not part of the
command to be entered)::
/data # luna -v
luna-base version v1.1.0 (release date 11-Oct-2024)
luna-base build date/time Oct 13 2024 22:06:18
Eigen library v3.4.0
sqlite v3.41.2
To be practically useful, one would also bind a local folder to a contain volume: e.g. if the tutorial s.lst
data exist in the current local folder. Locally:
cd ~/tutorial
cat s.lst
nsrr01 edfs/learn-nsrr01.edf edfs/learn-nsrr01-profusion.xml
nsrr02 edfs/learn-nsrr02.edf edfs/learn-nsrr02-profusion.xml
nsrr03 edfs/learn-nsrr03.edf edfs/learn-nsrr03-profusion.xml
${PWD} on Posix systems) to /data/ inside the container:
docker run --rm -it -v ${PWD}:/data remnrem/lunalite
/data # cat s.lst
nsrr01 edfs/learn-nsrr01.edf edfs/learn-nsrr01-profusion.xml
nsrr02 edfs/learn-nsrr02.edf edfs/learn-nsrr02-profusion.xml
nsrr03 edfs/learn-nsrr03.edf edfs/learn-nsrr03-profusion.xml
/data # luna s.lst 1 -s DESC
CMD #1: DESC
options: sig=*
EDF filename : edfs/learn-nsrr01.edf
ID : nsrr01
Clock time : 21.58.17 - 09.20.17
Duration : 11:22:00 40920 sec
# signals : 14
Signals : SaO2[1] PR[1] EEG_sec[125] ECG[250] EMG[125] EOG_L[50]
EOG_R[50] EEG[125] AIRFLOW[10] THOR_RES[10] ABDO_RES[10] POSITION[1]
LIGHT[1] OX_STAT[1]
Any files written to /data/ will be available on the host machine
after quitting the container. You can mount multiple volumes to
different points in the container (some of which can be readonly,
e.g. for the EDF data). See Docker documentation for details. Use
Ctrl-D to quit the Docker container (which will then be removed, as we
specified --rm).
Reference: Dockerfile
This github repository
contains the latest version of the Dockerfile, and the corresponding
luna.docker wrapper. Pre-built images are sent to the
`remnrem/luna Docker Hub page
for distribution.