Hypnograms
Commands to show and summarize elements of sleep macro architecture
These commands require staging annotations to be present in an annotation file, as described below.
| Command | Description |
|---|---|
HYPNO |
Sleep macro-architecture summaries |
STAGE |
Output sleep stages per epoch |
DYNAM |
Summarize epoch-level outputs by NREM cycles |
HYPNO
Estimates multiple summary statistics based on the hypnogram
HYPNO produces individual-level summary statistics (e.g. total sleep
time, percent in each sleep stage, etc), epoch-level output
(e.g. cumulative elapsed sleep duration, etc), statistics on bout
lengths and stage transition counts/probabilities. NREM sleep cycles
are calculated based on a modified heuristic following Feinberg and
Floyd. Optionally, this
command also 1) edits the stage data prior to summarizing it, for
example, trimming leading wake, 2) emits annotations corresponding
to key hypnogram features that subsequent commands can use.
Below are the key tables of parameters and outputs
Definitions
This schematic illustrates some of the main hypnogram summaries:
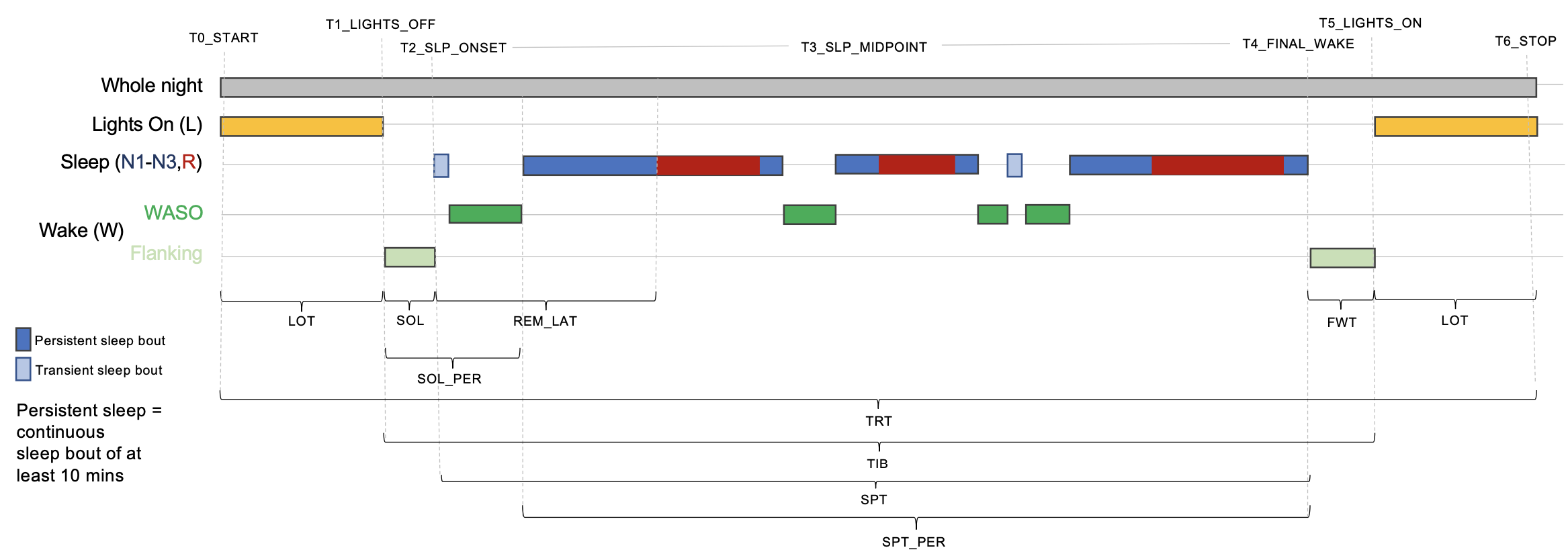
The key time markers (T0 to T6, which by definition are always in
this order) are:
T0: start of recordingT1: lights offT2: sleep onsetT3: sleep midpointT4: final wakeT5: lights onT6: end of recording
The primary spanning intervals are defined by these time points. The
entire recording (based on the epoched EDF) constitutes the total
recording time (TRT), from T0 to T6. If lights out/on markers are
present (see below), then time in bed (TIB) is the period between
lights out and lights on (T1 to T5); otherwise, it is the same as
TRT. Sleep period time (SPT) is from sleep onset to final wake (T2
to T4).
Sleep onset latency (SOL) is the time from lights out to sleep onset
(from T1 to T2). We further define persistent sleep as a sleep bout
of at least 10 minutes, and persistent sleep onset latency
(SOL_PER) is defined with regard to the first epoch of persistent sleep.
Persistent sleep onset latency
This is the first epoch in a
bout of at least 10 minutes, i.e. and not after ten minutes of
sleep. That is, if lights off is 10pm and the individual starts
a sleep bout at 10:25pm that lasts for 2 hours, then SOL_PER is
25 minutes - the same as SOL in this example - and not 25 +
10 = 35 minutes.
Wake is classified as wake after sleep onset (WASO) if it occurs between
sleep onset (T2) and final wake (T4). Wake outside the sleep period
(i.e. before sleep onset or after final wake) is called flanking
wake. Leading flanking wake duration is SOL (sleep onset
latency); we also record the duration of trailing flanking wake as
final wake time (FWT), i.e. up until lights on or the end of the recording.
Total wake time (TWT) is therefore SOL plus WASO plus FWT.
We also track the duration of lights on time (LOT, the period from T0
to T1 plus from T5 to T6).
Lights Off/Lights On
Any epoch with the stage annotation L means that lights are on,
i.e. wake but not part of the recording, and so not included as part
of TWT or SOL, etc. Lights on information can be encoded in
thgree ways, through epoch annotations, interval annotations, or
directly as a command argument.
Annotation encoding of lights off/on
In the first case, if an epoch/stage annotation is
L (i.e. rather than N1, N2, N3, R, W or ?), this implies
Lights On for that period. In the second case, interval annotations (or command options)
can specify either the time(s) at which a lights on or lights off events occurs,
or the interval which is to be considered as lights on or lights off. Given these
times/intervals, Luna will implicitly set the epoch encoding to L for all epochs
that are defined as lights on.
Interval encoding of lights on/off
Lights off and Lights on times will be derived from the
annotations, if available, as follows, based on the canonical
lights_on and lights_off annotations. These may represent
either change-points (i.e. lights_off has 0-seconds duration,
and just marks the exact time when the lights were turned off), or
an interval (i.e. lights_off has a non-zero durationm, implying
that it spans the entire period during which the lights were off).
The graphic below illustrates the logic Luna employs when trying
to derive the lights on/off times:
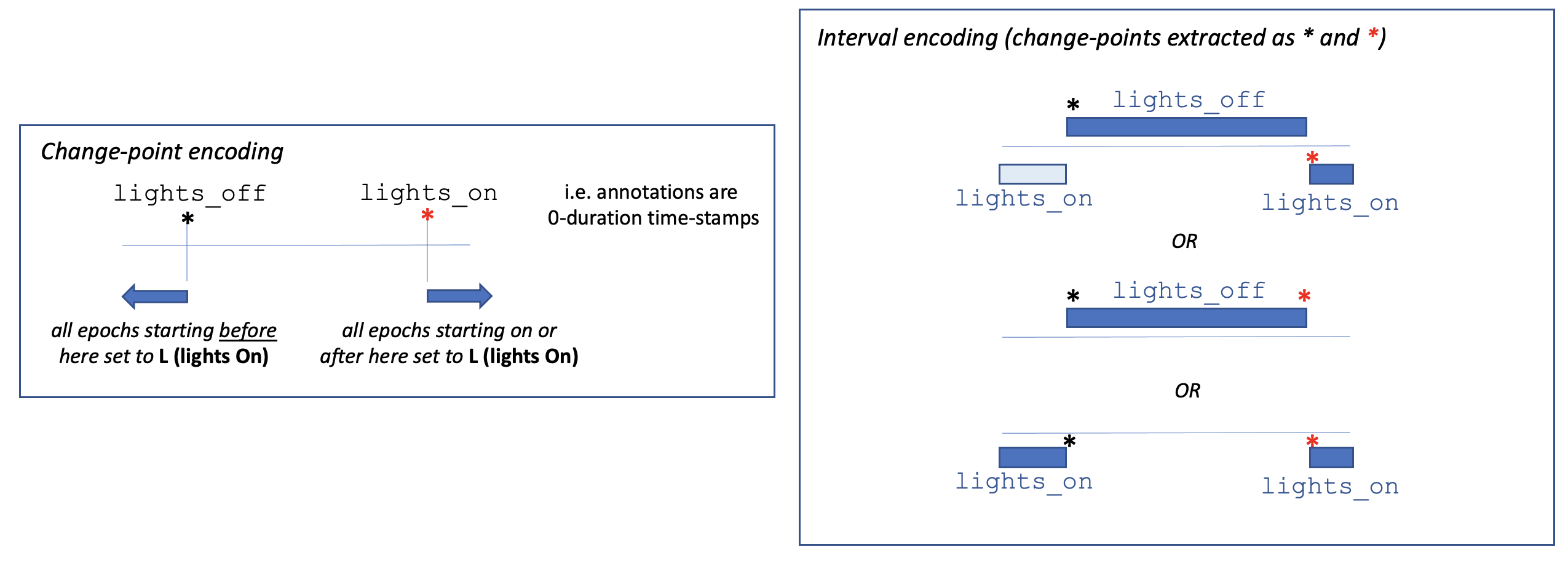
To summarize, lights off/on times (events) are extracted from interval annotations as follows:
| Valid combination | Off | On |
|---|---|---|
one lights_off interval only (with durtion at least one epoch) |
start of lights_off |
end of lights_off |
one lights_off interval only (with diration shorter than one epoch) |
start of lights_off |
end of recoding (i.e. no explicit lights_on) |
two lights_on intervals |
end of first lights_on |
start of second lights_on |
one lights_off + 1 or 2 lights_on |
start of lights_off |
start of last lights_on |
Note that the final case also works if two change-point events
(i.e. 0-duration intervals) are specified as lights_off and
lights_on). Having set a lights_off event: all epochs that end
before this time will be set to L. Having set a lights_on
event: all epochs that start on or after this time will be set to
L
If there are no Lights On epochs (L), this implies TRT equals
TIB and LOT equals 0, i.e. that Lights Off occurs at the start
of the recording and Lights On is at the end. Any leading/trailing
unscored epochs (?) are set to L (Lights On epochs), i.e if they
occur before/after the first/last sleep or wake epoch. This may not be
appropriate, however, e.g. if the recording continued for a long time
after the subject woke (as an example, see the first individual in the
tutorial dataset). Because of this, Luna implements
two versions of sleep efficiency: in both cases, the numerator is
total sleep time (TST); in the first case (SE), the denominator is
TIB, whereas for SME, the denominator is SPT. The latter metric
is often more robust is lights off/on has not been accurately tracked.
Explcitly setting lights off/on times
If lights off/on annotations are known by not present in the
annotations, they can be set explcitly via the lights-off and/or
lights-on options for the HYPNO command:
luna s.lst -o out.db -s HYPNO lights-off=22:30 lights-on=07:45
s.lst contained three individuals,
and times.txt was a tab-delimited text file:
ID LOFF LON
id1 22:00 07:00
id2 21:20 08:22:02
id3 . 09:00
. indicates it is unknown), this information can be included as follows:
luna s.lst -o out.db vars=times.txt -s HYPNO lights-off=${LOFF} lights-on=${LON}
Note that the variables ${LOFF} and ${LON} can be named
arbitarily, e.g. ${a} and ${b}. Also, lights off/on times can be
set as elapsed seconds from the EDF start rather than hh:mm or
hh:mm:ss (24-hour clock) values.
NSRR datasets are lights on/off
Many NSRR datasets do not
have explicit annotations that indicate when lights off/on events
occurred. Alternatively, lights off/on times can be set via
additional options to HYPNO, i.e. if they are known but not
explicitly represented in the staging annotations. (For example,
this is the case in the MrOS cohort, where lights on/off times are
available as part of the CSV datasets that accompany the signal
data.)
Excessive WASO
Some studies (e.g. if from home-based recordings) may contain
excessively long periods of leading or trailing wake. In a few
extreme cases in NSRR data, for example, a single N1 epoch may occur
hours before or after the main sleep period. By accepting this
outlier sleep epoch as real, this may lead to excessive estimates of
WASO (wake after sleep onset) or other parameters.
Especially in the context of home-based recordings where lights off/on
information might not have been explicitly specified, it may be more
appropriate to not treat this as defining the sleep period. In this
case, Luna implements the following heuristic, to set short outlier
periods of sleep to missing (?), if (by default) the cumulative
sleep period is no longer than five minutes, but is separated by at
least two hours (120 mins) of wake (either at the start or the end of
the recording). This schematic illustrates this process (note, here
using a 60 minute wake period, but as noted, Luna's default is 120
mins):

If you do not wish this heuristic to be used, set end-wake=0 and
end-sleep=0; alternatively, use these options to set other values.
Stage/epoch alignment
Typically, staging epochs will typically be typically 30 seconds and will be aligned with the signal data, i.e. starting at the start of the recording (0 seconds elapsed), as indicated in the top row in the picture below. In other cases, staging information may come in different formats. For example, even if scored in 30-second intervals, stage annotations may span longer intervals (multiples of 30 seconds, e.g. 90-seconds, 120-seconds, etc, to represent consecutive epochs of the same stage. Alternatively, the stage information may be represented as change-point events: i.e. 0-duration markers that indicate when a new stage starts (which is assumed to hold up until the next stage annotation). Both these alternatives will be automatically handled by Luna.

However, the fourth row represents a more problematic scenario: where
epochs do not align with Luna's internal epoch structure, which by
default starts at the start of the signal data (i.e. 0 seconds). This
will likely result in multiple epochs (from Luna's perspective) being
spanned by multiple, conflicting stage annotations. When this happens,
those epochs are set to ? and Luna issues a warning (and counts the
number of times this happens with the CONF variable in the output).
However, this can be easily avoided by telling Luna to alternatively start
encoding its own epochs so as to align with the external stage information, as per
the last row. See the EPOCH command for details. This can be accomplished as
luna s.lst -o out.db -s ' EPOCH align & HYPNO '
which means Luna will use the first instance of either N1, N2,
N3, R, W, ? or L to define where to start the first epoch.
(Note that after this, Luna will still assume that epochs will align,
i.e. that all stages are of similar duration, or multiples of that
duration, etc).
Trimming flanking wake
It is possible to trim leading/trailing flanking wake epochs via the
trim-wake (or trim-leading-wake and trim-trailing-wake) option.
These arguments take a value in minutes, and will set the
earlier/later flanking wake epochs to unknown (?) such that no more
than N minutes of flanking wake remains either before or after than
main sleep period.
Constrained analysis
The HYPNO command supports a range of constrained analyses of
hypnograms. This means that only a constrained portion of the
recording is used in analysis; internally, the remainder is set to
Lights On (L), i.e. it is effectively excluded from analysis.
Although beyond the scope of this documentation, this can be useful to calculate hypnogram statistics across different studies that may differ in the sleep opportunity period (i.e. so that some studies were effectively truncated). Alternatively, these approaches can be used to study ultradian dynamics in sleep macro-architecture.
The windows to analyze are generally defined by an anchor (by default sleep onset) and a duration (in minutes). For example:
-
consider only the first 240 minutes from sleep onset, by using the the
first=240option for which sleep onset is the default anchor (it can be set toanchor=T0oranchor=T1for recording start, lights out (or explicitly set toT2for sleep onset) -
consider only the last 240 minutes before sleep offset, by using the the
last=240option for which final wake is the default anchor (it can be set toanchor=T5oranchor=T6for lights on and recording end (or explicitly set toT4for final wake)
Alternatively, instead of specifying an anchor defined by the hypnogram, one can define it based on a fixed clock time: for example,
HYPNO first=60 clock=19:00:00
All outputs are as usual, with the exception of an additional SHORT
variable in the base strata if the window is larger than available (or
does not overlap at all, e.g. in clock mode, if the window ends before
the EDF start).
Interpretation of constrained hypnogram statistics
Obviously, certain metrics such as cycle-based analysis will be meaningless when applied to shorter
segements of a whole night of sleep. Many of these additional metrics can be skipped by adding
the verbose=T option, which will only output core baseline and stage-specific duration metrics.
Finally, one can specify a series of sliding windows to be automatically applied, via the slide option
which durations a window duration (in minutes) and a value to shift the window (also in minutes, always positive).
Instead of anchor, the slide command expects an anchor slide-anchor which can be 0 to 6, corresponding to
T0 to T6. For example: to have a two-hour window, shifted in 20 minute increments and starting at sleep onset (T2):
HYPNO slide=120,20 slide-anchor=2
slide-anchor=3, this uses sleep mid-point as the anchor
and shifts windows both forwards and backwards (until sleep onset, or final wake):
HYPNO slide=120,20 slide-anchor=3
Annotations
Adding the annot command generates a series of annotations (i.e. held
in memory, as if read in from an annotation file, and that can be used in MASK
commands or output with WRITE-ANNOTS) that capture key aspects of the hypnogram:
If annot=hyp the default prefix of h_ below is changed to hyp_.
Elapsed time
| Annotation root | Example(s) | Description |
|---|---|---|
h_clock |
h_clock_23, h_clock_00 |
Hour-intervals defined by clock-time |
h_elapsed_*_h* |
h_clock_N2_h1 |
Elapsed hour of stage, e.g. 1st hour of N2 |
h_elapsed_*_q* |
h_clock_S_q1 |
Quintile of elapsed stage, e.g. 1st quintile of sleep |
Elapsed hour (h_elapsed_*_h*) annotations are defined for: N1, N2, N3, NR (any
NREM), R, S (any sleep), W, WASO & T (total). Elapsed
quintile (h_elapsed_*_q*) annotations are defined for: N1, N2, N3, NR, R,
S, W & WASO.
Cycles
| Annotation root | Example(s) | Description |
|---|---|---|
h_cycle_n* |
h_cycle_n1, h_cycle_n2 |
Cycle number |
h_cycle_m* |
h_cycle_m0_10, h_cycle_m10_20 |
Intra-cycle period in minutes (10min bins) |
h_cycle_q* |
h_cycle_n1, h_cycle_n2 |
Intra-cycle period in quintiles (q1 to q5) |
Bouts
| Annotation root | Example(s) | Description |
|---|---|---|
h_bout05_* |
h_bout05_NR |
Within a bout of 5 or more minutes (W, R & NR only) |
h_bout10_* |
h_bout10_NR |
Within a bout of 10 or more minutes (W, R & NR only) |
Time-points
0-duration marker annotations:
| Annotation | Description |
|---|---|
h_t0_start |
Start of recording marker |
h_t1_lights_out |
Lights out marker |
h_t2_sleep_onset |
Sleep onset marker |
h_t3_sleep_midpoint |
Sleep midpoint marker |
h_t4_final_wake |
Final wake marker |
h_t5_lights_on |
Lights on marker |
h_t6_stop |
End of recording marker |
Transitions
0-duration marker annotations:
| Annotation | Description |
|---|---|
h_tr_NR_R |
NREM to REM transition |
h_tr_R_NR |
REM to NREM transition |
h_tr_NR_W |
NREM to wake transition |
h_tr_W_NR |
Wake to NREM transition |
h_tr_R_W |
REM to wake transition |
h_tr_W_R |
Wake to REM transition |
Epoch-level annotations
| Annotation | Description |
|---|---|
h_lights |
Epoch is Lights On |
h_waso |
Epoch is WASO |
h_persistent_sleep |
Epoch is in persistent sleep |
h_post_sleep_wake |
Epoch is post-sleep wake |
h_pre_sleep_wake |
Epoch is pre-sleep wake |
h_N2_asc |
Epoch is in ascending N2 |
h_N2_dsc |
Epoch is in descending N2 |
See below for examples of using a visualizing these annotations alongside a hypnogram.
Parameters
The primary options for HYPNO are tabulated here:
| Parameter | Example | Description |
|---|---|---|
epoch |
Display epoch-level output | |
annot |
hyp |
Add hypnogram-derived annotations, with prefix hyp_ (defaults to h_) |
lights-off |
23:30:00 |
Explicitly set Lights Off (hh:mm:ss or elapsed seconds from EDF start ) |
lights-on |
08:00:00 |
Explicitly set Lights On (hh:mm:ss or elapsed seconds from EDF start ) |
cache |
c1 |
Search cache for lights-off, lights-on markers (to work with EDGER |
end-wake |
120 | Threshold for trimming excessive WASO (mins) |
end-sleep |
5 | Threshold for trimming excessive WASO (mins) |
trim-wake |
20 | Trim leading/trailing wake to at most 20 mins |
trim-leading-wake |
20 | Trim leading wake to at most 20 mins |
trim-trailing-wake |
20 | Trim trailing wake to at most 20 mins |
Secondary/misc options
| Parameter | Example | Description |
|---|---|---|
file |
hypno.txt |
Optionally, read stage information from a file |
W |
waking |
Set the annotation class for wake epochs (instead of W) |
N1 |
Stg1 |
Set the annotaiton class for N1 epochs (if not N1) |
N2 |
Stg2 |
Set the annotaiton class for N2 epochs |
N3 |
Stg3 |
Set the annotaiton class for N3 epochs |
R |
RemSleep |
Set the annotaiton class for REM epochs |
? |
unk |
Set the annotation class for unscored/unknown epochs |
req-pre-post |
req-pre-post=5 |
For epoch-level transition flags, only consider post transition stage FLANKING_ALL values equal to or greater than this number of epochs (default 4) |
flanking-collapse-nrem |
flanking-collapse-nrem=F |
Collapse all NREM stages when considering flanking epoch similarity (default T) |
Constrained hypnogram options
| Parameter | Example | Description |
|---|---|---|
first |
240 | Only analyse the first 240 minutes for all statistics |
first-anchor |
T1 |
Anchor for extracting first N minutes (from T0, T1 or T2) |
By default, several common variants of stage labels are automatically
translated to the standard terms (N1, N2, N3, R, W, ? and
L), as tabulated below (including NSRR terms). If your values are
not listed here, you can use the options above (or
remap) to set them
manually.
| Stage/default | Aliases |
|---|---|
N1 |
NREM1, Stage1, Stage 1 sleep|1, S1, Stage1, SRO:Stage1Sleep, SDO:NonRapidEyeMovementSleep-N1 |
N2 |
NREM2, Stage2, Stage 2 sleep|2, S2, Stage2, SRO:Stage2Sleep, SDO:NonRapidEyeMovementSleep-N2 , NR |
N3 |
NREM3, Stage3, Stage 3 sleep|3, S3, Stage3, SRO:Stage3Sleep, SDO:NonRapidEyeMovementSleep-N3 |
N4 |
NREM4, Stage4, Stage 4 sleep|4, S4, Stage4, SRO:Stage4Sleep, SDO:NonRapidEyeMovementSleep-N4 |
R |
REM , SRO:RapidEyeMovement, SDO:RapidEyeMovementSleep, REM sleep|5 |
W |
wake, Wake|0 , SDO:WakeState, SRO:Wake |
L |
lights, lights_on |
? |
missing, unknown, artifact, A |
Stage translations
Note internally Luna can represent stage NREM4 sleep, as well as
Movement (M) and ambiguous/unscorable (U) epochs (as
distinct from missing/unknown/unscored epochs, ?). However,
for almost all purposes, Luna commands will automatically
translate NREM4 to N3; likewise, M and U epochs will be
treated as ?.
Outputs
The tables below show the various summary statistics based on this partitioning of sleep macro architecture, and are grouped as follows:
- individual level durations
- efficiency & latency metrics
- flags for unusual/corrupt staging
- sleep fragmentation indices
- stage-specific metrics
- epoch-level metrics
- cycle-level metrics
- stage transitions metrics
Individual-level durations (all metrics in minutes) (strata: none)
| Variable | Description |
|---|---|
TRT |
Total Recording Time: based on all scoring epochs (T0 – T6) |
TIB |
Time In Bed: Lights Off to Lights On (mins) (T1 – T5) |
SPT |
Sleep period time: sleep onset to final wake (T2 – T4) |
SPT_PER |
Persistent sleep period time: persistent sleep onset to final wake |
TST |
Total sleep time |
TST_PER |
Total persistent sleep time |
TWT |
Total wake time during Lights Off = SOL + WASO + FWT |
WASO |
Wake time between sleep onset and final wake onset (T2 – T4) |
FWT |
Duration of wake from final wake onset to Lights on (T4 – T5) |
PRE |
Time pre-sleep (relative to EDF start, versus lights off (SOL) |
POST |
Time post-sleep (relative to EDF stop) |
LOT |
Lights On duration (mins) |
TGT |
Total gap time (mins) - applicable to discontinuous EDFs |
Efficiency/latency metrics (strata: none)
| Variable | Description |
|---|---|
SE |
Sleep efficiency, TST / TIB (denom. = T1 – T5) |
SME |
Sleep maintenance efficiency, TST / SPT (denom. = T2 – T4) |
SOL |
Sleep latency (T1 – T2) |
SOL_PER |
Persistent sleep latency (T1 to onset of persistent sleep) |
REM_LAT |
REM latency (sleep onset T2 – first REM epoch) |
REM_LAT2 |
REM latency excluding WASO, i.e. elapsed NR at REM onset |
As noted above, there are two versions of sleep efficiency: in both
cases, the numerator is total sleep time (TST); in the first case
(SE), the denominator is TIB, whereas for SME, the denominator
is SPT. The latter metric is often more robust is lights off/on has
not been accurately tracked.
Luna also estimates REM latency (REM_LAT) as the time from sleep onset until
the first epoch of REM sleep. A second version of REM latency only counts
the sleep time between sleep onset and REM onset, i.e. ignoring any intervening
wake (REM_LAT2).
Indicators of unusual/corrupt staging (strata: none)
| Variable | Description |
|---|---|
FIXED_LIGHTS |
Number of epochs set to L before/after lights out/on |
LOST |
Lights On sleep duration (mins) : sleep set to L (should be 0) |
FIXED_WAKE |
Wake epochs set to L due to extreme WASO intervals |
FIXED_SLEEP |
Sleep epochs set to L due to extreme WASO intervals |
CONF |
Number of epochs w/ conflicting stages (should be 0) |
SINS |
Recording starts in sleep (0=N, 1=Y) |
EINS |
Recording ends in sleep (0=N, 1=Y) |
OTHR |
Duration of non-sleep/non-wake annotations (unknown/movement) |
Sleep fragmentation indices
| Variable | Description |
|---|---|
SFI |
Sleep Fragmentation Index: sleep to W transition count / TST |
TI_S |
Stage Transition Index (excludes W) N1-N2-N3-R transition count / TST |
TI_S3 |
3-class Stage Transition Index, NREM-REM-wake transition count / SPT |
TI_RNR |
REM-NREM transition count / TST |
LZW |
LZW complexity index (5-stages) |
LZW3 |
LZW complexity index (3-stages: NR, R & W) |
Stage-level output (option: none , strata: SS)
The following metrics are calculated for the following categories:
N1, N2, N3, all NR, R, W, S, ?, L & WASO:
| Variable | Description |
|---|---|
MINS |
Stage duration (mins) |
PCT |
Stage duration as proportion of TST (N1, N2, N3, NR & R only) |
DENS |
Stage duration as a proportion of SPT (density) |
BOUT_N |
Number of bouts (contiguous stretches of this stage) |
BOUT_MD |
Median bout duration (mins) |
BOUT_MN |
Mean bout duration (mins) |
BOUT_MX |
Maximum bout duration (mins) |
BOUT_05 |
Stage duration, considering only bouts of at least 5 minutes (mins) |
BOUT_10 |
Stage duration, considering only bouts of at least 10 minutes (mins) |
In addition, HYPNO distinguishes ascending vs. descending (vs. “flat”) N2 for MINS & PCT based on a heuristic:
-
N2_ASC: ascending N2 : N3 → N2 → N1/R/ W -
N2_DSC: descending N2 : N1/R/W → N2 → N3 -
N2_FLT: "flat" N2 : ambiguous, e.g. flanked by REM on both sides, or mixtures
Window-level stage-specific output output (option: slide , strata: SS x W x MINS)
The following metrics are calculated for the following categories:
N1, N2, N3, all NR, R, W, S and WASO:
| Variable | Description |
|---|---|
DENS |
Stage duration as a proportion of window duration (density) |
Epoch-level output (option: epoch, strata: E)
| Variable | Description |
|---|---|
CLOCK_HOURS |
Start time of epoch (hours since midnight) |
CLOCK_TIME |
Start time of epoch (hh:mm:ss) |
MINS |
Start time of epoch (minutes since start of EDF) |
START_SEC |
Start time of epochs (seconds since start of EDF) |
STAGE |
Text description of sleep stage |
STAGE_N |
Numeric encoding of sleep stage (see STAGE example, above) |
OSTAGE |
Original stage (i.e. before any editting to set L) |
PERSISTENT_SLEEP |
Flag to indicate persistent sleep (more than 10 minutes of sleep has elapsed) |
PRE |
Flag to indicate a pre-sleep epoch |
SPT |
Flag to indicate epochs within the sleep period time |
WASO |
Flag to indicate wake after sleep onset |
POST |
Flag to idicate a post-sleep epoch |
E_N1 |
Cumulative elapsed N1 sleep (minutes) |
E_N2 |
Cumulative elapsed N2 sleep (minutes) |
E_N3 |
Cumulative elapsed N3 sleep (minutes) |
E_REM |
Cumulative elapsed REM (minutes) |
E_SLEEP |
Cumulative elapsed sleep (minutes) |
E_WAKE |
Cumulative elapsed wake (minutes) |
E_WASO |
Cumulative elapsed WASO (minutes) |
PCT_E_N1 |
Cumulative elapsed N1 as proportion of total N1 sleep |
PCT_E_N2 |
Cumulative elapsed N2 as proportion of total N2 sleep |
PCT_E_N3 |
Cumulative elapsed N3 as proportion of total N3 sleep |
PCT_E_REM |
Cumulative elapsed REM as proportion of total REM sleep |
PCT_E_SLEEP |
Cumulative elapsed sleep as proportion of total sleep |
FLANKING |
The minimum number of similarly-staged epochs going either forwards or backwards |
FLANKING_ALL |
The total number of similar epochs in this stretch of similar epochs |
N2_WGT |
Score to indicate ascending versus descending N2 sleep (see below) |
NEAREST_WAKE |
Number of epochs (forward or backwards) since nearest wake epoch (see below) |
TR_NR2R |
Number of epochs from this NREM epoch to a REM transition (see below) |
TOT_NR2R |
Total number of contiguous NREM epochs followed by REM (see below) |
TR_NR2W |
Number of epochs from this NREM epoch to a wake transition (see below) |
TOT_NR2W |
Total number of contiguous NREM epochs followed by wake (see below) |
| ... etc ... | Similar TR_ and TOT_ variables defined for other transitions |
CYCLE |
Cycle number, if this epoch is in a sleep cycle |
CYCLE_POS_ABS |
Absolute position of this epoch in the current NREM cycle (mins) |
CYCLE_POS_REL |
Relative position of this epoch in the current NREM cycle (0-1) |
PERIOD |
Cycle period: NREMP or REMP, or missing if not in a cycle |
AFTER_GAP |
Flag to indicate if this epoch comes after a gap (EDF+D) |
Information on each NREM sleep cycle (strata: C)
| Variable | Description |
|---|---|
NREMC_START |
First epoch number of this NREM cycle |
NREMC_MINS |
Total duration of this cycle (mins) |
NREMC_NREM_MINS |
Duration of NREM in this cycle (mins) |
NREMC_REM_MINS |
Duration of REM in this cycle (mins) |
NREMC_OTHER_MINS |
Minutes of wake and unscored epochs |
Transition probabilities (strata: PRE x POST)
| Variable | Description |
|---|---|
N |
Count of transitions |
P |
Joint probability |
P_PRE_COND_POST |
Conditional probability P( pre | post ) |
P_POST_COND_PRE |
Conditional probability P( post | pre ) |
Example
Here we run HYPNO on nsrr02 from the tutorial data:
luna s.lst nsrr02 -o out.db -s HYPNO epoch
The baseline, individual-level output for HYPNO contains summaries
such as total sleep time (TST). Here we use Luna's behead utility
to view the output in a more human-readable fashion:
destrat out.db +HYPNO | behead
ID nsrr02
CONF 0
E0_START 0
E1_LIGHTS_OFF 0
E2_SLEEP_ONSET 45.5
E3_SLEEP_MIDPOINT 260.5
E4_FINAL_WAKE 475.5
E5_LIGHTS_ON 597.5
E6_STOP 597.5
EINS 0
FIXED_LIGHTS 0
FIXED_SLEEP 0
FIXED_WAKE 0
FWT 122
HMS0_START 21:18:06
HMS1_LIGHTS_OFF 21:18:06
HMS2_SLEEP_ONSET 22:03:36
HMS3_SLEEP_MIDPOINT 01:38:36
HMS4_FINAL_WAKE 05:13:36
HMS5_LIGHTS_ON 07:15:35
HMS6_STOP 07:15:35
LOST 0
LOT 0
LZW 0.107329842931937
LZW3 0.0636998254799302
NREMC 5
NREMC_MINS 77.1
OTHR 0
POST 121.5
PRE 45.5
REM_LAT 43.5
REM_LAT2 31
SE 59.8326359832636
SFI 0.0503496503496504
SINS 0
SME 83.1395348837209
SOL 45.5
SOL_PER 63
SPT 430
SPT_PER 412.5
T0_START 21.3016666666667
T1_LIGHTS_OFF 21.3016666666667
T2_SLEEP_ONSET 22.06
T3_SLEEP_MIDPOINT 25.6433333333333
T4_FINAL_WAKE 29.2266666666667
T5_LIGHTS_ON 31.26
T6_STOP 31.26
TGT 0
TIB 597.5
TI_RNR 0.0643356643356643
TI_S 0.26013986013986
TI_S3 0.137209302325581
TRT 597.5
TST 357.5
TST_PER 321
TWT 240
WASO 72.5
As indicated above, this individual had five sleep cycles (NREMC).
We can view details about each cycle by querying the C stratum of
out.db:
destrat out.db +HYPNO -r C
ID C NREMC_MINS NREMC_NREM_MINS NREMC_OTHER_MINS NREMC_REM_MINS NREMC_START
nsrr02 1 72.5 34 12.5 26 92
nsrr02 2 133 118.5 5 9.5 237
nsrr02 3 42 40 2 0 503
nsrr02 4 105.5 72 9 24.5 675
nsrr02 5 32.5 31 1.5 0 887
Finally, we can view epoch-level data (here just a few rows, for a subset of the available variables):
destrat out.db +HYPNO -r E
ID E CLOCK_TIME E_SLEEP STAGE
... skipped ...
nsrr02 89 22:02:06 0 W
nsrr02 90 22:02:36 0 W
nsrr02 91 22:03:06 0 W
nsrr02 92 22:03:36 0 N2
nsrr02 93 22:04:06 0.5 N2
nsrr02 94 22:04:36 1 W
nsrr02 95 22:05:06 1 W
nsrr02 96 22:05:36 1 W
nsrr02 97 22:06:06 1 W
nsrr02 98 22:06:36 1 N2
nsrr02 99 22:07:06 1.5 N2
nsrr02 100 22:07:36 2 N2
nsrr02 101 22:08:06 2.5 N2
nsrr02 102 22:08:36 3 N2
nsrr02 103 22:09:06 3.5 W
nsrr02 104 22:09:36 3.5 W
nsrr02 105 22:10:06 3.5 W
nsrr02 106 22:10:36 3.5 W
... cont'd ...
Time encoding: Several variables use an hours since midnight 24-hour encoding, so that 20.5, for example, means 8:30pm, etc.
Stage nomenclature: We use the newer format of N1, N2, and N3
sleep for variable names, although if NREM4 sleep has been scored it
will be reported as NREM (although, for "consistency" the variable
name is N4 rather than NREM4).
Ascending and descending N2: Ascending N2 sleep is defined as N2
sleep that follows N3 and precedes N1, Wake or REM. In contrast,
descending N2 sleep follows wake, N1 or REM sleep and precedes N3
sleep. The epoch-level N2_WGT variable (which is only defined for
N2 sleep) is an index ranging from -1 to +1 to quantify this.
Specifically, it is calculated by looking at the first k non-N2
epochs that come before and after that N2 epoch. (By default, k is
10.) N3 epochs are scored +1 if they are prior to this epoch,
otherwise -1. Wake, N1 and REM epochs are scored -1 if they come
before this epoch, otherwise +1. The final N2_WGT score is the
average of these scores. Epochs that are, e.g., less than -0.5 might
be called descending, whereas epochs that are greater than +0.5
might be called ascending. Ascending and descending N2 epochs
are also flagged by annotations h_N2_asc and h_N2_asc from annot.
Transition and flanking epoch defintions: The epoch-level variables TR_NR2R, TR_NR2W, etc, coumt the number
of epochs of NREM sleep remaining until a transition to some other
state (REM or wake respectively). Similar variables are defined for
transitions from REM and wake. The corresponding TOT_ variables
indicate the total number of NREM epochs in that segment - i.e. and so
can be used to select "stable" periods of some state of a minimum
duration prior to a transition to some other state. As noted above,
the HYPNO option req-pre-post option can be used to specify a
minimum duration of the post stage in order to trigger these flags;
by default this is 4 epochs (2 minutes). That is, is epoch 1227 below
was in fact back to N2 sleep, then the command would not have
flagged these earlier N2 epochs as being prior to wake.
E STAGE TOT_NR2R TOT_NR2W TR_NR2R TR_NR2W
...
1220 N2 0 22 0 5
1221 N2 0 22 0 4
1222 N2 0 22 0 3
1223 N2 0 22 0 2
1224 N2 0 22 0 1
1225 W 0 0 0 0
1226 W 0 0 0 0
1227 W 0 0 0 0
1228 W 0 0 0 0
1229 W 0 0 0 0
...
To illustrate the use of flanking similar epochs:
E FLANKING_ALL FLANKING STAGE
...
648 2 0 N2
649 2 0 N2
650 3 0 W
651 3 1 W
652 3 0 W
653 1 0 N2
654 1 0 W
655 15 0 N2
656 15 1 N2
657 15 2 N2
658 15 3 N2
659 15 4 N2
660 15 5 N2
661 15 6 N2
662 15 7 N2
663 15 6 N2
664 15 5 N2
665 15 4 N2
666 15 3 N2
667 15 2 N2
668 15 1 N2
669 15 0 N2
670 40 0 N3
671 40 1 N3
672 40 2 N3
...
i.e. in the above, we see a group of 15 N2 epochs (thus FLANKING_ALL
is 15 for those N2 epochs); the FLANKING essentially shows how
close each of those N2 epochs is to some other non-N2 epoch. Thus can
be used to select out epochs that fulfill certain criteria, i.e. if
one then wants to look at other epoch-level signal properties as a
function of stable stage and/or stage transitions. The above was run
with the option flanking-collapse-nrem=F which enforces a
distinction between N1, N2 and N3 epochs. The default is actually to
treat these all as similar (for the purpose of evaluating flanking
epochs only), i.e. without the above option, the output for these
epochs would instead be:
E FLANKING_ALL FLANKING STAGE
...
654 1 0 W
655 80 0 N2
656 80 1 N2
657 80 2 N2
658 80 3 N2
659 80 4 N2
660 80 5 N2
661 80 6 N2
662 80 7 N2
663 80 8 N2
664 80 9 N2
665 80 10 N2
666 80 11 N2
667 80 12 N2
668 80 13 N2
669 80 14 N2
670 80 15 N3
671 80 16 N3
672 80 17 N3
...
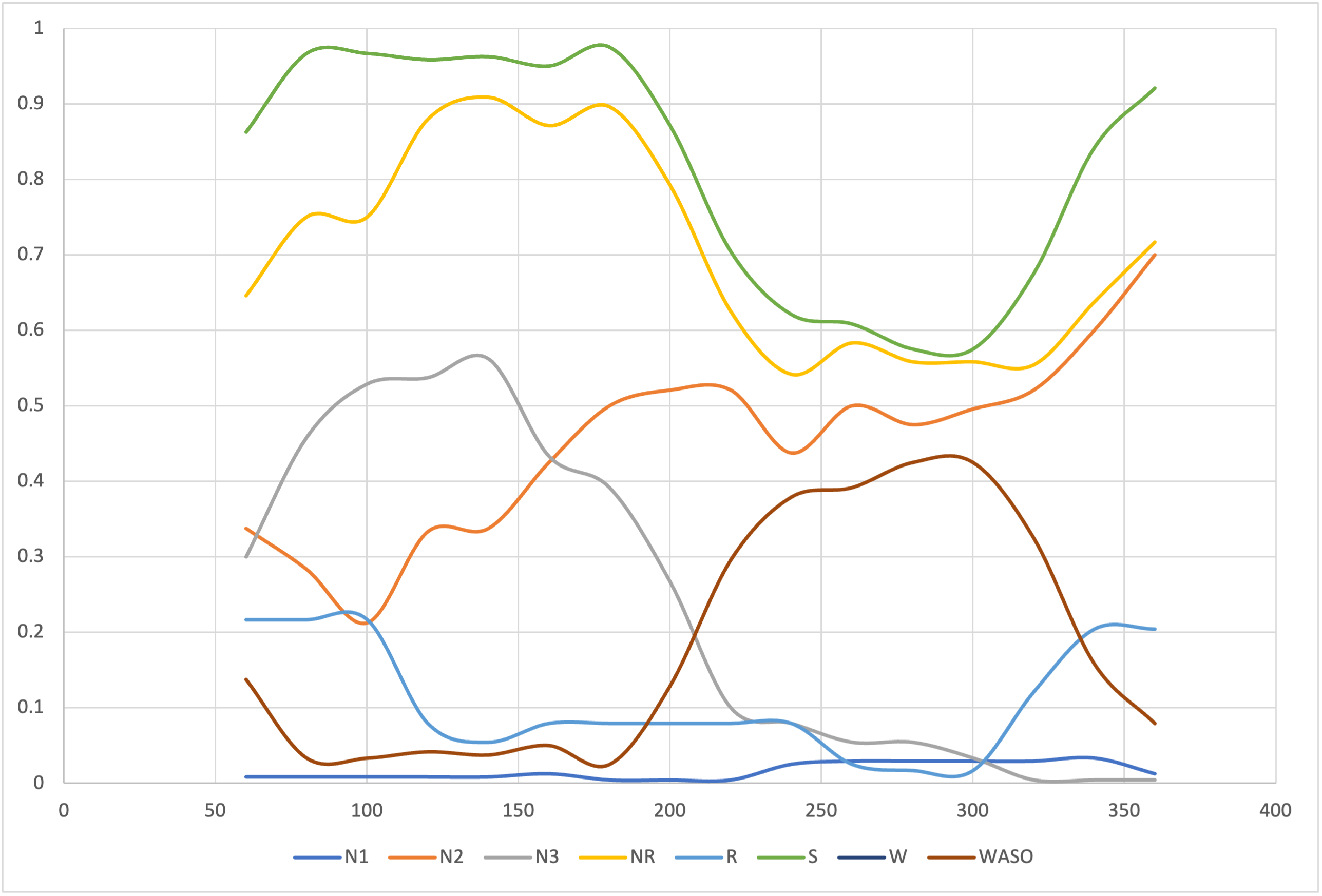
Constrained hypnogram statistics:
To give one example of constrained metrics - here stage density (DENS) for 2 hour windows, anchored on sleep onset
and incremented by 20 minutes each step:
luna s.lst -o out.db -s ' HYPNO slide=120,20 slide-anchor=2 verbose=F '
W, MINS and SS) - here omitting some columns for clarity, including ID and W:
destrat out.db +HYPNO -c SS -r W MINS -p 4
MINS DENS.SS_N1 DENS.SS_N2 DENS.SS_N3 DENS.SS_R DENS.SS_W
60 0.0083 0.3375 0.3000 0.2167 0.1375
80 0.0083 0.2833 0.4583 0.2167 0.0333
100 0.0083 0.2125 0.5292 0.2167 0.0333
120 0.0083 0.3333 0.5375 0.0792 0.0417
140 0.0083 0.3375 0.5625 0.0542 0.0375
160 0.0125 0.4250 0.4333 0.0792 0.0500
180 0.0042 0.5000 0.3917 0.0792 0.0250
200 0.0042 0.5208 0.2667 0.0792 0.1292
220 0.0042 0.5208 0.1000 0.0792 0.2958
240 0.0250 0.4375 0.0792 0.0792 0.3792
260 0.0292 0.5000 0.0542 0.0250 0.3917
280 0.0292 0.4750 0.0542 0.0167 0.4250
300 0.0292 0.4958 0.0333 0.0167 0.4250
320 0.0292 0.5208 0.0042 0.1208 0.3250
340 0.0333 0.6000 0.0042 0.2042 0.1583
360 0.0125 0.7000 0.0042 0.2042 0.0792
Plotting these values shows essentially a smoothed version of the hypnogram, but with metrics that can be compared between individuals.
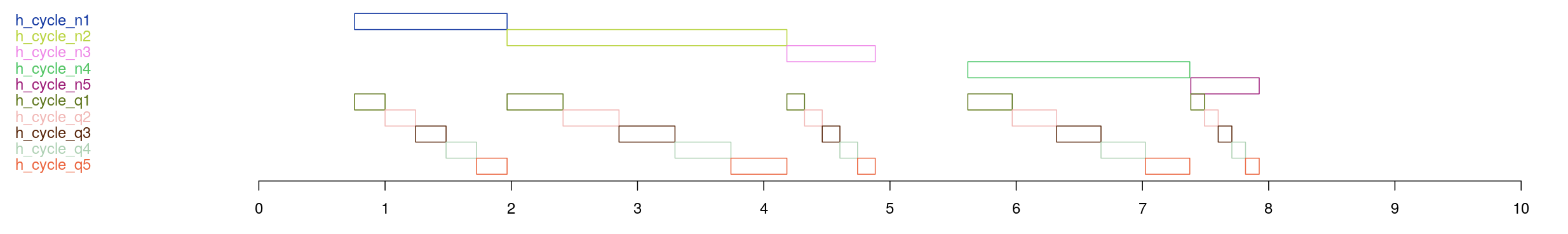
Annotations
To illustrate the types of annotations created by HYPNO annot, here we used Moonlight. Loading the Example
recording, under the Luna tab we entered the command: HYPNO annot. This adds a large number of annotations to the dataset.
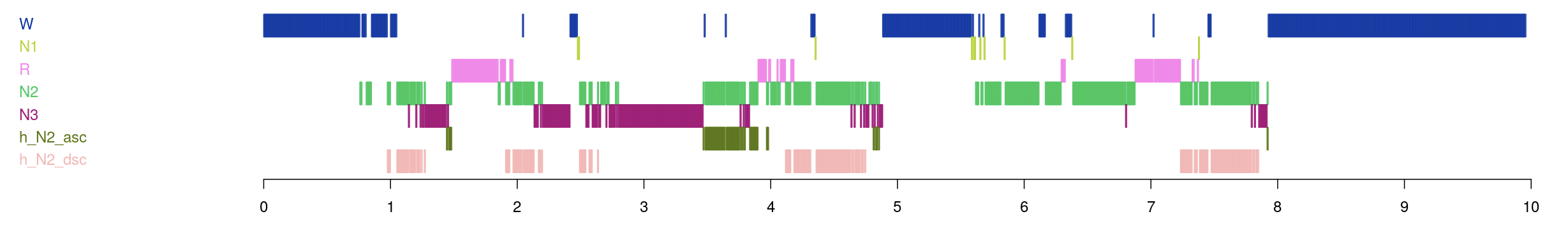
We then went to the Annotations tab and selected various ones to display. First, this is the overall hypnogram for this individual:

These annotations illustrated NREM cycles - the first five annotations track which of the five NREM cycles this individual has a particular epoch belong to. For example, if an epoch is spanned by the annotation h_cycle_n2, it belongs to this persons second NREM cycle. The second set of annotations show the relative positions within each NREM cycle, by quintile.
The next set illustrate ascending and descending N2 annotations, displayed against the original stage annotations:
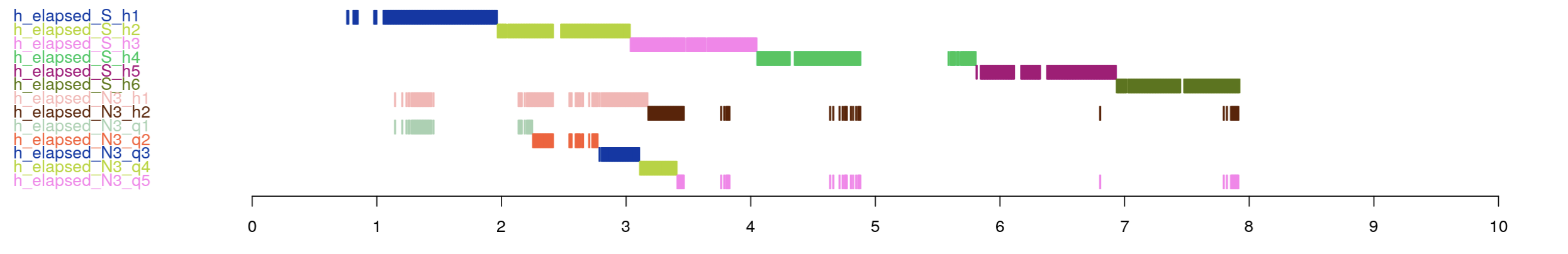
The final set show elapsed sleep (in hours), followed by elapsed N3 (also in hours), followed by elapsed N3 in quantiles.
The idea is that these annotations can then be used to select subsets of the night for particular analyses, e.g. cycle-specific analyses of spectral power, using something like the following:
HYPNO annot
FILTER sig=EEG bandpass=0.3,35 ripple=0.01,0.01 tw=0.5,5
uV sig=EEG
MASK ifnot=N2,N3
RE
ROBUST-NORM sig=EEG
CHEP-MASK sig=EEG ep-th=4,4,3
CHEP epochs
RE
FREEZE F1
TAG CYC/1
MASK ifnot=h_cycle_n1
PSD sig=EEG dB
FREEZE F1
TAG CYC/2
MASK ifnot=h_cycle_n2
PSD sig=EEG dB
FREEZE F1
TAG CYC/3
MASK ifnot=h_cycle_n3
PSD sig=EEG dB
FREEZE F1
TAG CYC/4
MASK ifnot=h_cycle_n4
PSD sig=EEG dB
FREEZE F1
TAG CYC/5
MASK ifnot=h_cycle_n5
PSD sig=EEG dB
FREEZE F1
TAG CYC/6
MASK ifnot=h_cycle_n6
PSD sig=EEG dB
For example, this shows the dissipation of delta power between successive NREM cycles, and an increaase in NREM beta:
destrat out.db +PSD -r CH CYC -c B/DELTA -v PSD -p 4
ID CH CYC PSD.B_DELTA
id1 EEG 1 5.3158
id1 EEG 2 4.5749
id1 EEG 3 -2.2887
id1 EEG 4 0.1554
id1 EEG 5 -3.7668
id1 EEG 6 -5.5107
STAGE
Export sleep stage information
This command simply reports sleep stage information (e.g. as encoded
stages in an annotation file.
Internally, it creates a single annotation class called SleepStage,
with instances that correspond to W, N1, N2, N3, R, ? and
L (Lights On). The HYPNO command does the same but
additionally computes a number of other statistics. Unlike
HYPNO however, the STAGES command can be run after epochs have
been masked out. In contrast, HYPNO requires the
original, entire EDF in order to produce meaningful summary
statistics on sleep macro architecture.
The canonical epoch labels for sleep/wake staging are as above
(i.e. N1, N2, N3, R, W as well as ? and L). Note that
alternatives such as NREM2, REM or wake are also acceptable (but
will be mapped to the above labels). Note that by default NREM4 is
mapped to N3 sleep.
Info
There is no explicit representation of movement or artifact
(i.e. set those to ? in terms of epoch-level staging) in the
STAGES or HYPNO commands. Still, you can always track that
information (e.g. and MASK epoch based on
them as generic annotations however.
Parameters
| Parameter | Example | Description |
|---|---|---|
W |
W=wake |
Set the annotation class for wake epochs |
N1 |
N1=NREM1 |
Set the annotaiton class for N1 epochs |
N2 |
N2=NREM2 |
Set the annotaiton class for N2 epochs |
N3 |
N3=NREM3 |
Set the annotaiton class for N3 epochs |
R |
R=REM |
Set the annotaiton class for REM epochs |
? |
?=UNKNOWN |
Set the annotation class for unscored/unknown epochs |
dump |
Write stage labels to standard out (minimal output ) | |
eannot |
s.txt |
Write stage labels to a file |
min |
Dump minimalistic stage labels to standard output stream |
Output
| Variable | Description |
|---|---|
CLOCK_TIME |
Clock time (hh:mm:ss) |
MINS |
Elapsed time from start of EDF (minutes) |
STAGE |
Sleep stage (text value) |
STAGE_N |
Numeric encoding of sleep stage |
Numeric stage encoding: The STAGE_N encoding is designed to help
with quickly plotting a hypnogram: W is 1, N1, N2 and
N3 are -1, -2 and -3 respectively; R is 0; ? is 2. As
such, something like the following R command can produce a
hypnogram-like representation of the night:
plot( d$E , d$STAGE_N )
See also
The lstages() function in
lunaR provides a quick way to run the STAGE command
for a single EDF, returning just a vector of stage names.
DYNAM
Summarize outputs from other commands (or inputs from a file) in terms of hypnogram-derived NREM cycle dynamics
This command will typically be invoked by adding dynam as an option
to one of the following commands that currently supports it: PSD,
COH, SPINDLES, SO, PSI, CORREL. Alternativey, epoch-level
inputs can be specifed from a file, in which case this functionality
is directly invoked via the DYNAM command. The same functions are
executed in either case.
This commands requires that HYPNO has previously been run, as it
relies on knowing the NREM cycle structure of a recording.