Summaries
Basic commands to give overviews of the structure of an EDF
| Command | Description |
|---|---|
DESC |
Simple description of an EDF, sent to the console |
SUMMARY |
More verbose description, sent to the console |
HEADERS |
Tabulate (channel-specific) EDF header information |
CONTAINS |
Indicate whether certain signals/stages/annotations are present |
ALIASES |
Display aliases assigned for channels and annotations |
TYPES |
Display current channel types |
VARS |
Display current individual-level variables |
TAG |
Generic command to add a tag (level/factor) to the output |
STATS |
Basic signal statistics (min/max, mean, RMS, etc) |
SIGSTATS |
Hjorth parameters and other signal statistics |
TABULATE |
Tabulate discrete values in a signal |
DUPES |
Finds flat signals and digital duplicates |
DESC
Basic information on the attached EDF, written to the console
Writes the EDF filename, ID, EDF duration, number of signals and the labels and sampling rates of those signals. If multiple EDFs are specified in a sample list, this information will be repeated for each one.
Parameters
| Parameter | Example | Description |
|---|---|---|
channels |
channels |
Only output simple list of channel names, one per row |
Outputs
Text written to the log/console. Note that, unlike most Luna
commands, the DESC command does not generate any other output, i.e. via
Luna's formal output mechanism.
Example
Using DESC on the command-line with a single EDF:
luna my.edf -s DESC
EDF filename : my.edf
ID : id001
Clock time : 23:07:56 - 07:39:00
Duration : 08:31:04
# signals : 6
Signals : EOG-L[256] EOG-R[256] EMG[256] EEG1[256] EEG2[256] EEG3[256]
SUMMARY
A more verbose display of EDF header information, written to the console
Similar to DESC, this command writes basic information from the EDF
header to the console; per-channel information from the
EDF header is also displayed.
Parameters
There are no command options for SUMMARY.
Outputs
Text written to the log/console. Note that, unlike most Luna
commands, SUMMARY does not generate any other output, i.e. via
Luna's formal output mechanism.
Example
To obtain a SUMMARY on the command-line with a single EDF:
luna my.edf -s SUMMARY
Patient ID : my.edf
Recording info :
Start date : 07.06.16
Start time : 23:07:56
# signals : 6
# records : 30664
Rec. dur. (s) : 1
Signal 1 : [EOG-L]
# samples per record : 256
transducer type : G02
physical dimension : uV
min/max (phys) : -125/125
EDF min/max (phys) : -125/125
min/max (digital) : -2048/2047
EDF min/max (digital): -2048/2047
pre-filtering : LP:35.00Hz HP:0.30Hz NOTCH:0
Signal 2 : [EOG-R]
# samples per record : 256
transducer type : G03
physical dimension : uV
min/max (phys) : -125/125
EDF min/max (phys) : -125/125
min/max (digital) : -2048/2047
EDF min/max (digital): -2048/2047
pre-filtering : LP:35.00Hz HP:0.30Hz NOTCH:0
... (etc) ...
HEADERS
Tabulate EDF header information
This command produces similar information to the SUMMARY command,
except it uses Luna's standard output
mechanism, rather than writing to the
console.
Parameters
| Parameter | Example | Description |
|---|---|---|
sig |
sig=${eeg} |
Only report HEADERS outputs for these channels |
signals |
Add a SIGNALS variable to the output, that lists all signals as a comma-delimited string |
Outputs
Basic EDF header information (strata: none)
| Variable | Description |
|---|---|
EDF_ID |
ID (in EDF header), or period (.) if empty |
EDF_TYPE |
EDF type (EDF, EDF+C or EDF+D) |
START_DATE |
Start date |
START_TIME |
Start time |
STOP_TIME |
Stop time |
NR |
Number of records |
NS |
Number of signals/channels (current in-memory EDF) |
NS_ALL |
NS in the original EDF file (before any manipulations) |
REC_DUR |
Duration of each record (seconds) |
TOT_DUR_SEC |
Total duration of EDF (seconds) |
TOT_DUR_HMS |
Total duration of EDF (hh:mm:ss string) |
SIGNALS |
(Optionally, if signals) A comma-delimited string of all channel labels |
Per-channel header information (strata: CH)
| Variable | Description |
|---|---|
POS |
Position (signal slot) in EDF |
DMAX |
Digital max |
DMIN |
Digital min |
PDIM |
Physical dimension (unit) field |
PMAX |
Physical min |
PMIN |
Physical max |
SR |
Sample rate (Hz) |
TRANS |
Transducer type field |
SENS |
Sensitivity (unit/bit) |
TYPE |
Inferred channel type |
Example
To obtain each channel's sampling rate from a single EDF my.edf, and create an output database out.db:
luna my.edf -o out.db -s HEADERS
out.db:
destrat out.db
--------------------------------------------------------------------------------
out.db: 1 command(s), 1 individual(s), 16 variable(s), 120 values
--------------------------------------------------------------------------------
command #1: c1 Fri Aug 14 12:38:29 2020 HEADERS sig=*
--------------------------------------------------------------------------------
distinct strata group(s):
commands : factors : levels : variables
-------------:-----------:--------------:---------------------------
[HEADERS] : . : 1 level(s) : EDF_ID EDF_TYPE NR NS NS_ALL REC_DUR
: : : START_DATE START_TIME STOP_TIME
: : : TOT_DUR_HMS TOT_DUR_SEC
: : :
[HEADERS] : CH : 14 level(s) : DMAX DMIN PDIM PMAX PMIN POS SENS
: : : SR TRANS TYPE
-------------:-----------:--------------:---------------------------
This shows there are two strata: the first is a baseline strata,
i.e. with factor . (period) to indicate no stratification, meaning
there is only one value for that variable for that EDF. The second
strata is by channel (CH), which has 14 levels, corresponding to
the 14 channels/signals in the EDF.
To extract the baseline information from the HEADERS command:
destrat out.db +HEADERS | behead
ID nsrr01
EDF_ID .
EDF_TYPE EDF
NR 40920
NS 14
NS_ALL 14
REC_DUR 1
START_DATE 01.01.85
START_TIME 21.58.17
STOP_TIME 09.20.16
TOT_DUR_HMS 11:22:00
TOT_DUR_SEC 40920
To extract only the sample rate variable (SR), which is stratified
per-channel (CH):
destrat out.db +HEADERS -r CH -v SR
ID CH SR
nsrr01 SaO2 1
nsrr01 PR 1
nsrr01 EEG(sec) 125
nsrr01 ECG 250
nsrr01 EMG 125
nsrr01 EOG(L) 50
nsrr01 EOG(R) 50
nsrr01 EEG 125
nsrr01 AIRFLOW 10
nsrr01 THOR RES 10
nsrr01 ABDO RES 10
nsrr01 POSITION 1
nsrr01 LIGHT 1
nsrr01 OX STAT 1
Alternatively, to extract all variables for EMG and ECG channels only:
destrat out.db +HEADERS -r CH/EMG,ECG
ID CH DMAX DMIN PDIM PMAX PMIN POS SENS SR TRANS TYPE
nsrr01 ECG 127 -128 mV 1.25 -1.25 4 0.00980 250 . ECG
nsrr01 EMG 127 -128 uV 31.5 -31.5 5 0.24705 125 . EMG
Note, when adding signals it can be useful to also set the special
variable order-signals=T on the Luna command line, so that
e.g. B,C,A is listed the same as A,B,C (e.g. when enumerating the
number of unique channel combinations that exist across different
EDFs).
CONTAINS
Indicate whether particular signals, annotations or staging are present
This command can be used to check whether certain necessary attributes (e.g. a particular signal or annotation, or the presence of staging) that may be required for a particular analysis are present.
CONTAINS is unusual among Luna commands, in that it can be run in different modes:
-
reporting to the standard output mechanisms (as detailed below)
-
optionally, set a variable based on the tested conditions
-
optionally, skipping processing the current EDF if the conditions aren't met
-
optionally, using the exit code or return code to report its findings based on all individuals/EDFs tested
Return codes
Using the bash shell, the default return code (i.e. after
running any command, not just Luna) is 0, meaning "success". It can
be accessed via the bash $? special shell variable.
For checking signals and annotations, the following convention is used:
- 0 : all signals present (in all individuals)
- 1 : at least 1 signal present (in all individuals)
- 2 : no signals present (in at least one individual)
For stages,
- 0 : sleep stage annotations present (in all individuals)
- 1 : sleep stage annotations absent (in at least one individual)
Windows
We have not tested this on any Windows machines, but the DOS variable
%errorlevel% should be the analogue of the Linux/macOS $? bash variable.
Most shells other than bash support the $? variable, although there may be some
variations.
Parameters
This command can be run with either the sig or stages options:
| Parameter | Example | Description |
|---|---|---|
sig |
${eeg} |
Channels to be checked for presence/absence |
annot |
Instead of signals, indicate whether specific annotations are present | |
stages |
Instead of signals, indicate whether sleep stage annotations are present | |
skip |
Skip to the next EDF unless all of the requested signals/annotations are present | |
skip-if-none |
Skip to the next EDF if none of the requested signals/annotations are present | |
var |
x |
Set variable ${x} to T or F instead of setting the exit code |
Output
The primary output of CONTAINS is via the return code, as described
above (and see example below). In addition, when checking whether the
EDF contains signals, some additional output is sent to the standard
output mechanism.
Individual-level output (strata: none)
| Variable | Description |
|---|---|
NS_REQ |
Number of requested channels |
NS_OBS |
Number of requested channels observed in the EDF |
NS_TOT |
Total number of channels in the EDF |
Channel-level output (option: sig, strata: CH)
| Variable | Description |
|---|---|
PRESENT |
0/1 variable to indicate whether the requested signal is present |
Output
First checking which signals are present via the DESC command:
luna cfs.lst 1 -s DESC
Signals : C3[128] C4[128] M1[128] M2[128] LOC[128] ROC[128]
ECG2[256] ECG1[256] EMG1[256] EMG2[256] EMG3[256] L_Leg[64]
R_Leg[64] AIRFLOW[32] THOR_EFFORT[32] ABDO_EFFORT[32] SNORE[256] SUM[32]
POSITION[1] OX_STATUS[1] PULSE[1] SpO2[1] NASAL_PRES[64] PlethWV[128]
Light[512] HRate[512]
To test whether all/some of the following are present in an automated manner, using CONTAINS: here to test
for LOC, ROC, EOG-L and/or EOG-R:
luna cfs.lst 1 -o out.db -s CONTAINS sig=LOC,ROC,EOG-L,EOG-R
To see the return code in this scenario (which is always from the last command executed):
echo $?
1
We can also look at the more detailed output:
destrat out.db +CONTAINS
ID NS_OBS NS_REQ NS_TOT
cfs-visit5-800002 2 4 26
destrat out.db +CONTAINS -r CH
ID CH PRESENT
cfs-visit5-800002 LOC 1
cfs-visit5-800002 ROC 1
cfs-visit5-800002 EOG-L 0
cfs-visit5-800002 EOG-R 0
In practice, the CONTAINS command is likely only to be used in scripting: e.g.
luna s.lst silent=T -s 'CONTAINS sig=${eeg}'
HAS_EEG=$?
if [[ ${HAS_EEG} -eq 0 ]]; then
# ...
# ...EEG-specific code here...
# ...
fi
ALIASES
Output which annotation and channel remappings (aliases) were used for a particular individual
The alias and remap special options alter channel and annotation
labels on-the-fly. This command produces a record that tracks the
original labels and associated remappings.
Parameters
None
Output
Per-channel information (strata: CH)
| Variable | Description |
|---|---|
ORIG |
Original channel label (prior to re-aliasing) |
Per-annotation information (strata: ANNOT)
| Variable | Description |
|---|---|
ORIG |
Original annotation label (prior to re-mapping) |
Example
If the file vars.txt contains aliases for two EEG channels:
alias EEG1|EEG
alias EEG2|EEG(sec)
luna s.lst 1 @vars.txt -o out.db -s ALIASES
out.db:
commands : factors : levels : variables
----------------:-------------------:---------------:---------------------------
[ALIASES] : ANNOT : 5 level(s) : ORIG
: : :
[ALIASES] : CH : 2 level(s) : ORIG
: : :
----------------:-------------------:---------------:---------------------------
destrat out.db +ALIASES -r CH
ID CH ORIG
nsrr01 EEG1 EEG
nsrr01 EEG2 EEG(sec)
nsrr-remap=F flag is added):
destrat out.db +ALIASES -r ANNOT
ID ANNOT ORIG
nsrr01 apnea_obstructive Obstructive Apnea
nsrr01 arousal_standard Arousal ()
nsrr01 artifact_SpO2 SpO2 artifact
nsrr01 desat SpO2 desaturation
nsrr01 hypopnea Hypopnea
SpO2 artifact but these were (automatically) remapped to artifact_SpO2.
TYPES
Displays curret channel type definitions, either based
on the default set (internal to Luna) or user-specified (via
ch-match, ch-exact and/or ch-clear).
Types can be useful when writing scripts: e.g. to generate power spectra for all EEG channels:
PSD sig=${eeg} spectrum
Parameters
None
Output
A list of channel IDs and the associated type are listed to standard output. This command will give identical output for all EDFs (i.e. it is effectively independent of the attached EDF, although currently, it is necessary to supply an EDF). That is, this command shows the template used to assign channel type variables for any EDF.
Example
luna s.lst 1 -s TYPES
EXACT HRate HR
PARTIAL OFF IGNORE
PARTIAL STATUS IGNORE
PARTIAL E1 EOG
PARTIAL E2 EOG
PARTIAL EOG EOG
PARTIAL LOC EOG
PARTIAL ROC EOG
PARTIAL ECG ECG
PARTIAL EKG ECG
... etc ...
The complete internal table of type definitions is here.
Users can add types via one or more ch-match or ch-exact options, or
clear all default types with ch-clear=Y.
-
Exact matches are listed (and processed) before partial matches; they are case-sensitive, and require the full label to match.
-
Partial matches are processed after exact matches; these are case-insensitive, and the full channel label does not need to match: e.g. the template
O2will matchSpO2. -
Channels are assigned to types in a specific order, depending on the type. That is, the channel is assigned the first type that matches, all possible later matches are ignored. See here for details.
The ch-exact and ch-match options can take comma-delimited lists, with each element in the form TYPE|channel.
If multiple channels specified at once, one can write as element in the form TYPE|channel1|channel2, etc. For example:
luna s.lst ch-exact="EEG|xx|yy" -s TYPES
EXACT HRate HR
EXACT xx EEG
EXACT yy EEG
PARTIAL OFF IGNORE
PARTIAL STATUS IGNORE
PARTIAL E1 EOG
PARTIAL E2 EOG
...
VARS
Output all variables for an individual
The VARS command tabulates both
run-level and
individual-level for each
EDF/individual. The primary value of this command is to provide a
record of which values were used for a particular run/set of commands.
Parameters
None
Output
Per-variable information (strata: VAR)
| Variable | Description |
|---|---|
VAL |
Value for this variable, for this individual |
INDIV |
Boolean (0 or 1) to indicate whether this is an individual-level variable |
Example
Running VARS for one individual, setting the run-level variable ${xyz} on the command line:
luna s.lst 1 xyz=123 -o out.db -s VARS
VAR):
destrat out.db +VARS -r VAR
ID VAR INDIV VAL
nsrr01 apnea 0 apnea_obstructive,apnea_central,apnea_mixed,hypopnea
nsrr01 arousal 0 arousal_standard,arousal_spontaneous,arousal_external,arousal_respiratory,arousal_plm,arousal_cheshire
nsrr01 arrhythmia 0 bradycardia,tachycardia,tachycardia_narrowcomplex
nsrr01 artifact 0 artifact_respiratory,artifact_proximal_pH,artifact_distal_pH,artifact_blood_pressure,artifact_TcCO2,artifact_SpO2,artifact_EtCO2
nsrr01 n1 0 NREM1
nsrr01 n2 0 NREM2
nsrr01 n3 0 NREM3,NREM4
nsrr01 plm 0 plm_left,plm_right
nsrr01 rem 0 REM
nsrr01 sleep 0 NREM1,NREM2,NREM3,NREM4,REM
nsrr01 wake 0 wake
nsrr01 xyz 0 123
nsrr01 airflow 1 AIRFLOW
nsrr01 ecg 1 ECG
nsrr01 eeg 1 EEG(sec),EEG
nsrr01 effort 1 THOR_RES,ABDO_RES
nsrr01 emg 1 EMG
nsrr01 eog 1 EOG(L),EOG(R)
nsrr01 generic 1 NA
nsrr01 hr 1 PR
nsrr01 id 1 nsrr01
nsrr01 ignore 1 NA
nsrr01 leg 1 NA
nsrr01 light 1 LIGHT
nsrr01 oxygen 1 SaO2,OX_STAT
nsrr01 position 1 POSITION
nsrr01 snore 1 NA
We see the specified variable is present (with the value 123), and
with 0 under the INDIV column, denoting that this variable would
have been similarly defined for all EDFs present in this run (in this
particular example, there happens to be only a single EDF). We also a
number of other, automatically defined variables. The top ones are
automatically generated by Luna, to facilitate working with NSRR
annotations (e.g. the automatic variable ${apnea} which is defined
as apnea_obstructive,apnea_central,apnea_mixed,hypopnea). We also
see some individual level automatic variables, defined according to
channel types, e.g. ${airflow}.
Note that in the above output NA means not available, i.e. that variable was not
defined for that run/individual. Although displayed as NA by
destrat, the actual value would be blank (e.g. if ${snore} was in a
command file, it would be replaced by a blank, 0-length character
string, rather than an actual NA character string).
TAG
Used to mark specific analyses in output
TAGs allow you to arbitrarily add extra levels by which output is
stratified, which can be useful to distinguish similar commands
performed within the same analysis-run (for example, if several rounds
of MASKs and RESTRUCTUREs
are specified in one analysis).
Parameters
| Parameter | Example | Description |
|---|---|---|
| {tag} | run/L1 |
Add tag with level L1 to factor run in output |
tag |
tag=run/L1 |
Identical to the above, but explicitly using the tag option |
If a . (period) is specified as the level, that particular TAG is
removed from any subsequent output. For example, the following would
remove the run tag from all subsequent output:
TAG run/.
To remove all tags, use the following:
TAG .
Hint
Do not select a TAG factor name that is already
used by a Luna command (e.g. F, CH, E, B, etc). One safe way
to ensure this is by only using lower-case values for TAG factors,
as all internal factors are upper-case.
Output
No specific output is generated, beyond adding the specified stratifying factors in the output.
Example
By way of context, here we review the concept of strata that Luna
uses extensively in its output. For example, some analyses may have
their output stratified by two factors: channel and spectral band,
for which it uses the labels CH and B. The B (spectral power
band) factor has a number of levels, e.g. SIGMA and so on. When
using the destrat tool, one might request
output for all channels but only the sigma band, for example:
destrat out.db +PSD -r CH B/SIGMA
where the syntax is {factor}/{level} for spectral power. How does
this apply to tags? Consider the following command
script cmd.txt, which runs the
STATS command (described below):
% Create statistics for the entire night, tagged by ALL for the RUN factor
TAG run/ALL
STATS epoch
% Perform some kind of operation to change the data
EPOCH
MASK epoch=1-10
RESTRUCTURE
% If repeating the same STATS command, we need to distinguish
% the output from the prior STATS command, so we now set the 'run'
% factor to FLT, i.e. to indicate these are filtered results
% (Note that the choice of 'run', 'ALL' and 'FLT' are completely
% arbitrary)
TAG run/FLT
STATS epoch
If we run this command:
luna s.lst -o out.db < cmd.txt
and look at the output:
destrat out.db
we will see that the run factor (specified by the TAG command)
appears as an additional factor:
[STATS] : CH run : 28 level(s) : MAX MEAN MEDIAN MEAN_MD MEDIAN_MD
: : : RMS_MD SKEW_MD MIN NE NE1
: : : RMS SKEW
: : :
[STATS] : E CH run : (...) : MAX MEAN MEDIAN MIN RMS SKEW
If we, for example, want to extract the RMS variable, here only for
the channel EEG:
destrat out.db +STATS -v RMS -r CH/EEG run
then we'll see it has values for two levels of run, i.e. for ALL
and FLT, in other words before and after applying the mask:
ID CH run RMS
nsrr01 EEG ALL 37.801350632511
nsrr01 EEG FLT 17.0591938957608
Alternatively, to extract RMS for all channels but from only the
second set of values, one would write (mirroring the original TAG
run/FLT command, and adding -p 2 to control the number of decimal
places output):
destrat out.db +STATS -v RMS -r CH run/FLT -p 2
which yields:
ID CH run RMS
nsrr01 SaO2 FLT 95.24
nsrr01 PR FLT 73.18
nsrr01 EEG(sec) FLT 14.76
nsrr01 ECG FLT 0.07
nsrr01 EMG FLT 8.73
nsrr01 EOG(L) FLT 53.26
nsrr01 EOG(R) FLT 50.11
nsrr01 EEG FLT 17.06
nsrr01 AIRFLOW FLT 0.12
nsrr01 THOR_RES FLT 0.18
nsrr01 ABDO_RES FLT 0.06
nsrr01 POSITION FLT 2.01
nsrr01 LIGHT FLT 1.00
nsrr01 OX_STAT FLT 0.26
STATS
Calculates basic (per-epoch) statistics per-channel
Calculates the mean, median, RMS, standard deviation and min/max for
each channel. By default, this is for the entire duration of the
recording, i.e. including both masked and unmasked epochs. For epoched
data, adding the epoch option generates additional epoch-level
output as well as the median (across epochs) of the per-epoch mean,
median, RMS and skewness. Results from the epoch option are based
on unmasked epochs only.
Parameters
| Parameter | Example | Description |
|---|---|---|
sig |
sig=C3,F3 |
Restrict analysis to these channels |
epoch |
epoch |
Calculate per-epoch statistics |
pct |
F |
Output percentiles (1st, 5th, 10th, etc) ( default: T) |
min |
Only output the mean (minimal output ) |
Outputs
Whole-night, per-channel statistics, based on all epochs (strata: CH)
| Variable | Description |
|---|---|
MIN |
Signal minimum (from data, not EDF header) |
MAX |
Signal maximum (from data, not EDF header) |
MEAN |
Signal mean |
MEDIAN |
Signal median |
RMS |
Signal root mean square |
SKEW |
Signal skewness |
P01, P02, P05, ..., P99 |
Percentile values (unless pct=F) |
Per-epoch, per-channel statistics for unmasked epochs only (option: epoch, strata: CH x E)
| Variable | Description |
|---|---|
MIN |
Signal minimum (from data, not EDF header) |
MAX |
Signal maximum (from data, not EDF header) |
MEAN |
Signal mean |
MEDIAN |
Signal median |
RMS |
Signal root mean square |
SKEW |
Signal skewness |
Additional whole-night, per-channel statistics, only using unmasked epochs (option: epoch, strata: CH)
| Variable | Description |
|---|---|
NE |
Total number of epochs in record |
NE1 |
Number of unmasked epochs actually used in calculations |
MEAN_MD |
Median of all per-epoch means |
MEDIAN_MD |
Median of all per-epoch medians |
RMS_MD |
Median of all per-epoch RMS values |
SKEW_MD |
Median of all per-epoch skewness values |
Example
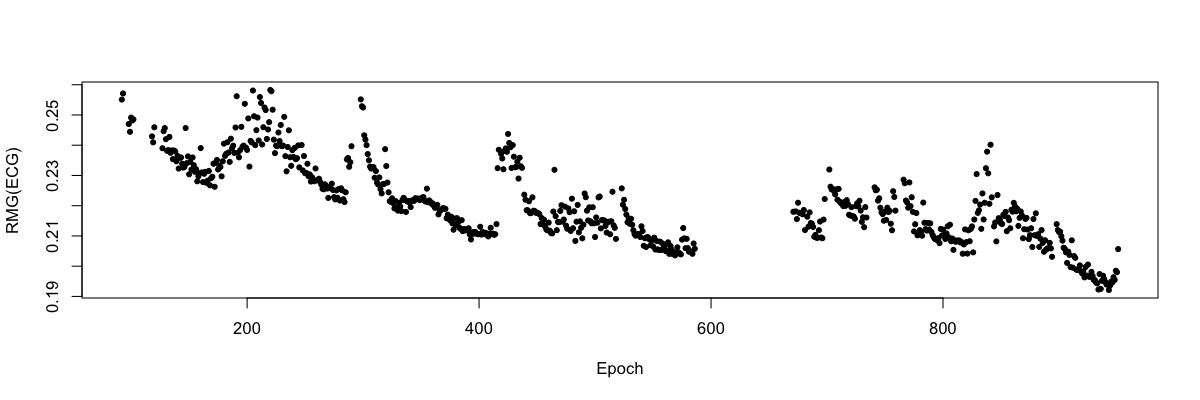
As an arbitrary example: to see how the RMS of the ECG signal changes over the night during sleep, using data from the tutorial data (second individual in this case):
luna s.lst nsrr02 -o out.db -s "EPOCH & MASK if=wake & RESTRUCTURE & STATS epoch sig=ECG"
destrat out.db +STATS -r E CH -v RMS > res.txt
Using the R package, load and plot these epoch-wise values (run these commands within R):
d <- read.table("res.txt",header=T)
plot( d$E , d$RMS , col = d$ID , pch=20 , xlab = "Epoch" , ylab = "RMG(ECG)" )
SIGSTATS
Epoch-wise Hjorth parameters and other statistics
This command calculates and reports per-epoch (and whole-signal) Hjorth parameters and (optionally) other statistics: signal root mean square (RMS), indices of signal clipping (the proportion of points that equal the minimum or maximum for that epoch), absolute maximum absolute values and flatness (proportion of points of a similar value to the preceding value).
Parameters
Core parameters:
| Parameter | Example | Description |
|---|---|---|
sig |
sig=C3,F3 |
Restrict analysis to these channels |
epooch |
Epoch-level output | |
sr-over |
100 |
Requires a sample rate of at least 100 Hz to report signal statistics |
Additional statistics
| Parameter | Description |
|---|---|
pfd |
Compute signal Petrosian fractal dimension |
pe |
Compute signal permutation entropy |
pe-m |
Embedding dimension for permutation entropy (default 3 to 7) |
pe-t |
Step function for permutation entropy (default 1) |
rms |
|
flat |
1e-6 |
clipped |
|
max |
100 |
Output
Per-channel whole-signal statistics (strata: CH)
| Variable | Description |
|---|---|
H1 |
First Hjorth parameter (activity) |
H2 |
Second Hjorth parameter (mobility) |
H3 |
Third Hjorth parameter (complexity) |
CLIP |
Proportion of clipped sample points |
MAX |
Proportion of maxed out sample points |
FLAT |
Proportion of flat sample points |
RMS |
Signal root mean square |
Per-channel epoch-level statistics (strata: CH x E)
| Variable | Description |
|---|---|
H1 |
First Hjorth parameter (activity) |
H2 |
Second Hjorth parameter (mobility) |
H3 |
Third Hjorth parameter (complexity) |
CLIP |
Proportion of clipped sample points |
MAX |
Proportion of maxed out sample points |
FLAT |
Proportion of flat sample points |
RMS |
Signal root mean square |
TABULATE
Tabulates discrete values in a signal
Most signals in EDFs are continuously-valued; for discrete signals
(e.g. body position encoded as an integer value, or a binary 0/1
status signal), the TABULATE command can provide useful summaries.
Parameters
| Parameter | Example | Description |
|---|---|---|
sig |
sig=position,status |
Restrict analysis to these channels |
req |
req=1000,5000 |
Count # of distinct values w/ at least this many (e.g. 1000 or 5000) observations |
Output
Per-channel tabulation statistics (strata: CH)
| Variable | Description |
|---|---|
NV |
Number of discrete values observed for this channel |
Per-channel tabulation statistics of required counts (option: req; strata: CH x REQ)
| Variable | Description |
|---|---|
NV |
Number of discrete values observed with at least REQ observations per value |
Per-channel/value tabulation statistics (strata: CH x VALUE)
| Variable | Description |
|---|---|
N |
Number of sample points for this value/channel |
Example
Here we have an EDF with a POSITION channel that encodes body position
via integer values in the EDF. We can use TABULATE to give a breakdown of the observed values:
luna s.lst -o out.db -s TABULATE sig=POSITION
NV to tell us how many unique values were observed:
destrat out.db +TABULATE -r CH
ID CH NV
id001 POSITION 4
i.e. there are four distinct values observed for this signal.
Info
Note, if the EDF had been masked and restructured, the observed values would reflect only the retained portion of the dataset.
To find out what those values are, we can look at the CH x VALUE strata of the output:
destrat out.db +TABULATE -r CH VALUE
ID CH VALUE N
id001 POSITION 0 1029
id001 POSITION 1 25515
id001 POSITION 2 944
id001 POSITION 3 13432
That is, the four values are 0, 1, 2 and 3. Note that they
can actually be any floating point values, i.e. they need not start at
0 or be integers, although the output of TABULATE is more likely
to be interpretable when the signal comprises a small set of integer
values. Also note that the 16-bit floating-point nature of EDF encoding
means that (depending on how the signal was encoded with respect to physical and digital min/max
values, integer values may in fact be encoded as floating point values numerically close
to the integer (e.g. 2.999 instead of 3).
The N variable here is the number of sample points observed -
i.e. it should be divided by the sample rate of the channel to get the
implied duration in seconds; in this case of a 1 Hz POSITION signal,
the values can be directly interpreted as seconds.
As a note, the HEADERS or SUMMARY options can also show the physical min/max in the EDF - e.g. here
we also see a range from 0 to 3 (although just because the EDF has a certain min/max range specified in
the header, this does not imply that the full range is necessarily observed in the actual signal):
Signal 12 : [POSITION]
sampling rate : 1 Hz
# samples per record : 1
transducer type :
physical dimension :
min/max (phys) : 0/3
EDF min/max (phys) : 0/3
min/max (digital) : 0/3
EDF min/max (digital): 0/3
pre-filtering :
As a simple convenience feature, we can limit observations to only those with at least a certain number of occurrences
with req option:
luna s.lst -o out.db -s 'TABULATE sig=POSITION req=500,1000,5000'
Looking at the CH x REQ output stratum, we see NV varies as a
function of REQ, i.e. only 2 values with over 5000 sample points
(nb. this also could have been trivially computed based on the CH x
VALUE outputs of course):
destrat out.db +TABULATE -r CH REQ
ID CH REQ NV
id001 POSITION 500 4
id001 POSITION 1000 3
id001 POSITION 5000 2
Finally, to interprete the data POSITION signal: if the values correspond to categories, we naturally need external
information to tell us what those values are. For example, we may know that the following mapping holds:
| Signal value | Mapping |
|---|---|
0 |
Left position |
1 |
Right position |
2 |
Supine position |
3 |
Front position |
For this type of signal, it can often be convenient to create a correspondoing interval-based annotation also,
which can be accomplished via the S2A (signal-to-annotation) command.
luna s.lst -s 'S2A sig=POSITION encoding=left,0,right,1,supine,2,prone,3 & WRITE-ANNOTS hms file=^.annot '
added 13 intervals for left based on -0.05 <= POSITION <= 0.05
added 28 intervals for prone based on 2.95 <= POSITION <= 3.05
added 30 intervals for right based on 0.95 <= POSITION <= 1.05
added 24 intervals for supine based on 1.95 <= POSITION <= 2.05
class instance channel start stop meta
start_hms 21.58.17 . . . .
duration_hms 11.22.00 . . . .
duration_sec 40920 . . . .
epoch_sec 30 . . . .
supine . POSITION 21:58:17 22:00:39 .
right . POSITION 22:00:39 22:00:40 .
prone . POSITION 22:00:40 22:00:42 .
right . POSITION 22:00:42 22:00:43 .
supine . POSITION 22:00:43 22:00:44 .
right . POSITION 22:00:44 22:00:45 .
supine . POSITION 22:00:45 22:00:49 .
prone . POSITION 22:00:49 22:00:50 .
...
DUPES
Finds digital/physical signal duplicates (and flat signals)
This function is designed to spot obviously redundant or empty channels in an EDF. It compares all pairs of channels (either on the digital or physical values in the EDF) and flags duplicates (as well as flat channels).
Comparisons are made with some epsilon value to define "different" (i.e. this could allow for small numerical differences in otherwise identical signals). As soon as the command detects as difference it stops evaluating that pair of channels.
Parameters
| Parameter | Example | Description |
|---|---|---|
sig |
sig=position,status |
Restrict analysis to these channels |
physical |
Use physical instead of digital values | |
eps |
0.001 | Set epsilon value (default 0.01) |
prop |
0.2 | Percentage of epoch that have to differ for something to be called "different" (default 0.1) |
Output
Per-individual statistics (strata: none)
| Variable | Description |
|---|---|
INVALID |
Number of invalid channels (no non-zero physical range) |
DUPES |
Number of duplicate channel pairs |
FLAT |
Number of flat channels |
Per-channel statistics (strata: CH)
| Variable | Description |
|---|---|
DUPE |
Indicator (0/1) of whether this channel is a duplicate with some other |
FLAT |
Indicator (0/1) of whether this channel is flat |
Per-channel-pair statistics (strata: CHS)
| Variable | Description |
|---|---|
DUPE |
Value (1) set if this pair of channels (A,B) is a duplicate |
Example
See the Luna walk-through for an example.