Interval annotations
Here we describe the formats of annotation files, how to attach them to EDFs, and how to view and summarize their contents.
| Command | Description |
|---|---|
| Luna annotations | Overview of annotations in Luna |
| .annot files | .annot file format (the preferred Luna annotation format) |
| .eannot files | .eannot file format |
| NSRR XML files | NSRR XML annotation files |
--xml & --xml2 |
View NSRR XML annotation files |
REMAP |
Apply annotation remappings after loading |
ANNOTS |
Tabulate all annotations |
MAKE-ANNOTS |
Make new annotations |
WRITE-ANNOTS |
Write annotations as .annot or .xml |
META |
Add meta-data to annotations |
SPANNING |
Report on coverage of annotations |
ESPAN |
Epoch-based annotation coverage |
A2S |
Add a 0/1 signal based on an annotation |
S2A |
Add an annotation based on ranged values of a signal |
ALIGN-ANNOTS |
Align epochs (paired with ALIGN-EPOCHS) |
Luna annotations
Annotations are a key part of Luna, and in combination with the
MASK command and so-called eval
expressions, provide a flexible way to manipulate EDFs.
Luna can represent a number of different types of annotation. Annotations may be represented as arbitrary intervals of time (defined with respect to the start of the EDF), or can be specified at the per-epoch level in a number of ways. First, we introduce some terminology:
A class of annotations is a set of arbitrary time intervals (e.g. such as "spindles")
Each instance of a class (e.g. one particular spindle) is defined by a time interval and an identifier (which may or may not be unique).
Optionally, each instance can also have associated meta-data, stored as typed key/value pairs.
This table summarizes these levels of annotation:
| Type | Level | Example concept | Specific example |
|---|---|---|---|
| class name | Generic set of annotations | All spindles | spindles |
| instance | Specific instance of a class defined by a time interval |
A single spindle | 873.2 to 874.1 seconds |
| instance names | ID (or non-unique label) for each instance | Spindle number (or type of spindle) |
spindle-1, spindle-2, ... (or fast-spindle, slow-spindle) |
| instance meta data | Associated information | Duration, amplitude, etc, for an individual spindle |
dur=0.88 amp=12.2 frq=11.1 |
Annotations can be represented in a number of different file formats, all described below:
.annotfiles, where each row describes an event/interval and any associated meta-data (including times, channels, etc).eannotfiles containing simple per-epoch labels, where each distinct label is treated as a distinction annotation class- EDF+ Annotations, where annotations are embedded in the EDF as separate channels
- Luna-format XML files
- NSRR-format XML files, where each ScoredEvent parent node in one XML is treated as a distinct annotation class
Note
Currently masks only operate on class and instance level
data, not meta-data. That is, annotation meta-data is currently
not used by Luna.
Also, including or excluding certain annotations with the
annot option works at the class level
only. That is, either all instances of a given class are
loaded in, or none are.
.annot files
Generic annotation files
This is the preferred (and most flexible) annotation format for generic event/interval-based annotations. These are text-based, tab-delimited files that containing interval-level annotations.
Note
.annot files should be plain-text but need not have the
.annot file extension. Any annotation file that is neither
.xml nor .eannot is assumed to be of generic .annot
file format.
An .annot file can describe one or more annotations classes. Each
class can have one or more instances, where each instance
corresponds to an interval of time and a single row of the .annot
file.
The standard .annot specification involves at least six
tab-delimited fields (optionally allowing whitespace delimiters
instead). There are numerous options and alternatives the supplement
this basic format, as outlined below:
- Column formats : full (six-column) versus reduced (3 or 4 column) formats
- Headers : optional rows prior to data rows
- Time-encoding specifications : different ways to specify the start/stop times of intervals/events
Columns
The standard definition specifies at least six fields, as described below.
The full (internal) representation of annotation data as is follows:
| Column | Description | Default |
|---|---|---|
class |
Annotation class | Required |
instance |
Instance ID | If not used, specify . |
channel |
Channel(s) | If not applicable, specify . |
start |
Start time | Required: see time-encoding specifications below |
stop |
Stop time | See time-encoding specifications below |
meta |
Meta-data | Values or key-value pairs; if not present, specfiy . |
This six-column format is the recommended format, and is what is
written (by default) by WRITE-ANNOTS.
Extra fields: any additional fields (seven onwards) are interpreted as additional
meta-data values, i.e. conceptually the same as the key=value pairs in the sixth
meta column, but allowing for a different format, as described below.
Reduced forms: For convenience, Luna also allows reduced 3-column and 4-column variant formats that skip certain columns. If only four columns are present, they must be as follows:
| Column | Description | Default |
|---|---|---|
class |
Annotation class | Required |
instance |
Instance ID | If not used, specify . |
start |
Start time | Required: see time-encoding specifications below |
stop |
Stop time | See time-encoding specifications below |
If only three columns are present, they must be as follows:
| Column | Description | Default |
|---|---|---|
class |
Annotation class | Required |
start |
Start time | Required: see time-encoding specifications below |
stop |
Stop time | See time-encoding specifications below |
At its 3-column minimal, the .annot format is simply an annotation class and start/stop times: e.g.
annot1 10.20 12.50
In this example, this data-rows specifies an annotation of class
annot1 that starts at 10.2 seconds (elapsed time from EDF start) and
lasts 2.3 seconds, ending at 12.5 seconds.
Technically, a single annotation file can contain different rows with
different variants of these 6/4/3 column formats: we do not advise
this, however, as it means that the file will not be easily readable
by tools such as R that expect regular formats (although a regular
version can be easily generated with WRITE-ANNOTS).
Rows with 1, 2 or 5 columns will be reported as errors. Further, if there is a header field that specifies additional tabular meta-data columns (i.e. columns 7 onwards), then all rows must have the same number of fields as specified in the header.
Headers
Optionally, .annot files can have multiple header rows (the preamble) that define the
classes in that file. Each of these header rows starts with a # character and
contains between one to three |-delimited fields, which are class
name, description and meta-data types respectively. For example
# a1
# a2 | Unlike the first, this annotation has a description field
# a3 | This one also specifies meta-data types | val1[txt] val2[num] val3[bool]
In the contrived example above, there are three annotation classes:
a1, a2 and a3. Only a2 and a3 have description fields. If
an annotation class includes meta-data (i.e. a3), it
must be first defined in a header row.
Optionally, .annot files can also contain a second type of header row,
prior to any data-row, which specifies and labels the subsequent columns.
If this row exists, it must exactly conform to one of the following
(which also acts as an internal check on the ordering of columns and
general correctness of the file):
For the six-column format:
class instance channel start stop meta
class instance start stop
class start stop
If including optional tabular meta-data fields (columns 7 onwards), those labels can be
any unique valid key labels (e.g. no special characters or spaces, etc): for example,
here we add three extra meta-data tags with keys AMP, dur_sec and F1:
class instance channel start stop meta AMP dur_sec F1
As noted above, It is advised that subsequent data rows adopt the same column format.
This then allows R (or other packages) to read the .annot file as a
tab/whitespace-delimited table, with the columns given these variable
names, e.g. using the read.table() function. As, by default,
read.table() ignores comment lines beginning with #, this will
skip the preamble:
d <- read.table( "file.annot" , header=T )
Time encoding
The .annot file recognizes various ways to specify start and stop
times for each annotation. These formats can be mixed-and-matched
within a single .annot file.
-
Elapsed seconds: this is the default - any numerical value is interpreted as seconds elapsed relative from the EDF start time (i.e. as specified in the EDF header)
-
Clock-time: any times with the format hh:mm:ss or hh.mm.ss are assumed to be 24-hour clock-times. These can include fractions of a second, e.g.
23:03:01.524(which is also the same as23.03.01.524, i.e. if the EDF.character is used to delimit hours, minutes and seconds insted of the colon (:) character). It is also possible to specify dates as well as clock-times, i.e. as is necessary for long recordings, see below -
Elapsed hh:mm:ss: any time starting
0+is assumed to be an elapsed time specified in hh:mm:ss format rather than a clock-time, e.g.0+00:00:30,0+00:01:00corresponds to 30 and 60 seconds past the EDF start -
Epoch-encoding: instead of elapsed or clock times, it is also possible to specify start and stop times in terms of epochs by starting entries with the
echaracter: see below for more details
Further, stop times can take one of two additonal encodings:
- Durations: if the stop time starts with
+it is interpreted as the duration of the interval, rather than the end. This can be used for any type of start time (elapsed seconds or clock-time). i.e.
annot1 10.00 +5
annot1 10.00 15.00
- Until next annotation: if the stop time is an ellipsis (
...), this means that this annotation spans until the start of the next annotation listed in that file (or the end of the EDF, if this is the last row). Naturally, this assumes the annotation in the next row starts after the start of the current row. This can be convenient if only changes in sleep stage are specified at irregular intervals: i.e.is the same asannot1 10.00 ... annot1 15.00 ... annot2 22.00 30.30annot1 10.00 15.00 annot1 15.00 22.00 annot2 22.00 30.00
Order of annotations
The order of annotations is generally irrelevant in .annot
files. (The one exception is when using the ... stop-time encoding,
which relies on the next row in that same file, irrespective of what
annotation class it is, or whether there is another intervening
annotation elsewhere in the file).
Luna can read from multiple annotation files for a given individual; further, the same annotation classes can be present in more than one file. Internally, all annotations will be merged into a single set of annotations, irrespective of which file they were read from.
Interval encoding
Intervals are defined to be inclusive of the start but exclusive of
the stop, i.e. the interval from a to b is [a,b). In other
words, b is the first point just past the end of the interval. Interval
duration is therefore defined as b-a. This means that two 30-second
epochs specified below are non-overlapping epochs: rather, they are
contiguous despite 30.00 appearing in both definitions:
class1 interval1 0.00 30.00
class1 interval2 30.00 60.00
Epoch encoding
Epoch encoding is provided as a convenience feature as many
annotations are in fact specified in terms of regular-sized epochs and
it might be awkward to always have to list the start and stop times of
each epoch. These codes (that start with the characters e: to
distinguish them from times in seconds) are converted to the
equivalent interval when reading the file.
If the value in the start or stop column begins with the characters
e:, then Luna assumes that epoch-notation is being used to specify the
interval. By default, Luna assumes non-overlapping 30-second epochs,
whereby e:1, e:2, e:3, etc, refer to the first, second, third,
etc, epochs. If the stop column also starts e: then Luna assumes the interval is
from the start for the first epoch to the end of the second epoch. Otherwise, the stop column
should be set to missing (.)
For example, the following three lines specify identical intervals:
class1 instance1 0 30
class1 instance1 e:1 .
class1 instance1 30 120
class1 instance1 e:2 e:4
Different epochs definitions can be specified by explicitly appending the epoch duration (in seconds) and increment (in seconds) as colon-delimited values, as shown in the Table below.
| Example | Description | Implied interval (sec) |
|---|---|---|
e:2 |
Second epoch; non-overlapping 30-second epochs | [30.0,60.0) |
e:2:20 |
Second epoch; non-overlapping 20-second epochs | [20.0,40.0) |
e:2:30:15 |
Second epoch; 50% overlapping 30-second epochs | [15.0,45.0) |
If not specified, the increment is assumed to be the same as the epoch duration, i.e. no overlap of consecutive epochs.
Note
As epochs are defined within the .annot file itself, the
actual EDF need not be epoched (i.e. from the
EPOCH command). In fact, the EDF may even
have epochs of a different duration specified. Also, unlike
.eannot files, not every epoch needs to be
specified in an .annot file, i.e. if there are 1200 epochs, you
do not need to have exactly 1200 rows in this file. This is
because instances specified with epoch-encoding are automatically
converted to interval-encoding upon loading.
Time resolution and floating-point accuracy
By default, Luna rounds all times read in annotation files to seconds with 4 decimal places. This level of temporal resolution (which should be sufficient for all PSG/EEG contexts) is imposed in order to avoid precision issues that are inherent in representing floating-point numbers digitally.
That is, unlike integers, many real number do not have an exact
internal floating-point representation: for example, 15.24 seconds may
internally be represented as 15.23333333333339 or (depending on the
platform or what prior conversions were done) 15.2400000000001, etc.
Internally, Luna uses a 64-bit integer time-track (in which each
time-point by default reflects 1e-9 seconds) to determine, e.g.,
whether intervals overlap or not. In both cases, 15.23333333333339
and 15.2400000000001 will therefore be mapped to the same time-point
up to 4 decimal places accuracy: i.e. 15240000000 rather than
15239999999, etc. This makes downstream handling of intervals more
robust.
Date-encoding
When using clock-time encoding, it is also possible to specify dates. The default
form uses European date format (following EDF specifications) and so expects
something in the form dd-mm-yy-hh:mm:ss, i.e. 31-12-99-23:59:59.
Delimiters: Alternatively, one can use / for the date, and/or a space instead of
- to separate the date and time, e.g. 31/12/99 23:59:59.
Month labels: it also also allowed to use the following
three-character codes (case-insensitive) instead of the numbers 1
through 12: Jan, Feb, Mar, Apr, May, Jun, Jul, Aug,
Sep, Oct, Nov and Dec.
M/D/Y and Y/M/D formats: By setting the special
variable date-format to either
MDY or YMD (versus the default value of DMY) you can instruct
Luna to assume these alternate date formats. Note, this implies that all annotation
files will have the same date format. All outputs (e.g. from WRITE-ANNOTS or HEADERS etc, will
always use European formats for dates, however. Although the EDF specification requires European dates
in the EDF header, if this is wrongly specified, you can use edf-date-format with the same convention.
Day offsets: Alternatively, you can use the special codes d1,
d2 etc in place of a full date to mean the first day, second day,
where day one is specified by the start date in the EDF header.
If a date is too far from the EDF start date (e.g. 100 days), the Luna will give an error. Annotations that start before the EDF start date will be ignored.
Data rows
Subsequent data rows of the .annot file specify instances of one
of these three classes, along with the necessary meta-data in the case
of a3. For example, in six-column format, the full file might read:
# a1
# a2 | Unlike the first, this annotation has a description field
# a3 | This one also specifies meta-data types | val1[txt] val2[num] val3[bool]
class instance channel start stop meta
a1 i1 . 10.00 15.00 .
a1 i2 . 92.10 105.22 .
a1 i3 . 108.5 123.11 .
a2 . . e:2 . .
a2 . . e:7 . .
a2 . . e:10 e:12 .
a3 A C3 0 +30 W;0.88;Y
a3 A C3,C4 30 +30 W;0.98;Y
a3 B C4 60 +30 N1;0.23;N
That is, each data row specifies one annotation of the previously
defined classes a1, a2, and a3.
- first column: class name matches one of the header rows (i.e.
a1,a2ora3in this example); if a new class is encountered, it is added on-the-fly - second column: instance ID that can be unique or not with respect to its annotation class (or even missing, as for
a2) - third column: channel IDs can be optinally specified, if they are relevant. e.g. for events detected on a given channel(s).
- fourth and fifth column(s): these define the interval for this annotation instance, as described above
- sixth column: either missing (period,
.) or, if the header specified meta-data for that annotation class (as fora3above), then these must be listed in a pipe-delimited format, in the same order as they were declared in the header/preamble.
As noted above, this example also uses different ways to specify start/stop times. You can mix and match these different formats with the same annotation
class. (In the example above, we map them to a1, a2, and a3 simply to make the example clearer.)
The same information could also be represented in a reduced format (i.e. without header rows or extra columns) but of course some information would be lost (e.g. meta-data and channels for a3, and instance IDs):
a1 10.00 15.00
a1 92.10 105.22
a1 108.5 123.11
a2 e:2 .
a2 e:7 .
a2 e:10 e:12
a3 0 +30
a3 30 +30
a3 60 +30
Meta-data
Meta-data are typed user-defined attributes of individual annotation
instances. For example, annotations for spindle events may have an
amplitude or frequency meta-data attribute. The .annot format
provides a few ways to specify annotation meta-data, in column six (or
beyond):
-
key-value pairs: this is the default used by
WRITE-ANNOTS, to specify meta-data askey=valuepairs in the sixthmetafield. -
values: if a full header has been specified with meta-data fields, then values in the sxith
metafield are assumed to correspond to those tags, as shown below -
tabular meta-data: given a header row that specifies columns beyond the sixth, values in those columns are treated as meta-data
That is, here are the three ways of specifying the same information: key-value pairs, which don't require any prior specification of the types:
a . . 10 20 v1=abc;v2=22
a . . 20 30 .
b . . 40 80 .
Stating the meta-fields in the extended header that describes annotation class a:
# a | annotation 'a' | v1[txt] v2[num]
...
a . . 10 20 abc;22
a . . 20 30 . (?? check)
b . . 40 80 .
Third, as tabular meta-data where the standard header row specifies the fields (which must be defined explicitly as missing for all annotation classes):
class instance channel start stop meta v1 v2
...
a . . 10 20 . abc 22
a . . 20 30 . . .
b . . 40 80 . . .
As illustrated above, in the original example the a3 class also expects some meta-data for
each instance: three variables named var1, var2 and var3, each with a specified type.
Meta-data types
The following types are currently available in Luna:
| Type | Description |
|---|---|
num |
Numeric (i.e. any floating point number) |
int |
Integer |
bool |
Boolean yes/no, true/false (with values y, yes, Y or 1 versus n, N, no or 0) |
txt |
Any text string |
Types can be defined on a file-by-file basis in the header rows, as described above.
You can also explicitly set meta-data keys to a given with the options
num-atype, int-atype, txt-atype and bool-atype, all of which
take a comma-delimited list of key labels. These apply to all files.
If no type if specified, all annotations are assumed to be
numeric. This behavior can be changed by setting the special variable
annot-meta-default-num to F.
Meta-data types primarily matter only for the DERIVE command currently.
Formats
Key-value meta-data should be delimited by either ; or | characters. e.g.
a . . 10 20 v1=abc;v2=22
a . . 20 30 v1=ced|v2=33
These defaults can be changed with the annot-meta-delim1 and annot-meta-delim2 special options.
The assignment symbol (which is = by default) can also be changed
(but has to be a single character): e.g. annot-keyval=: to allow
a . . 10 20 v1:abc;v2:22
a . . 20 30 v1:ced|v2:33
Note that these options apply to all annotations files read: if you
have a mixture of formats, you should use WRITE-ANNOTS first to make
a uniform set.
Standard format
The .annot file is designed to support a range of input formats, for
flexibility. As described below, no matter what the form of inputs,
any valid .annot file will be output in a more limited, standardized
manner by the WRITE-ANNOTS command, as described
below.
.eannot files
This is the simplest format for epoch-level annotations. Epoch
annotations in .eannot files are simple labels attached to
individual epochs. The format is as follows:
- one row per epoch
- each row contains a single label, that is attached to that epoch
- for each distinct label in the file, a new annotation class is generated
- each instance is assigned the same ID as the label name (i.e. same as the class name)
When an .eannot is specified in the
sample-list, it is attached prior
to loading the EDF. By default, Luna assumes epochs are 30-seconds in
duration and do not overlap when using .eannot files.
Hint
To work with .eannot files but use different epoch definitions, you have three options:
-
use the
EPOCHandEPOCH-ANNOTcommands to attach the file after initially attaching the EDF (i.e. instead of specifying the.eannotfile in the sample-list) -
Set the special variable
epoch-lenvariable if the.eannotis specified in the sample list (i.e. and so loaded when the EDF is first attached, prior to running anyEPOCHcommand) -
Use the
e:epoch encoding notation in a generic.annotfile instead, as described above.
Unlike other types of annotation, these can be loaded via a Luna
command, EPOCH-ANNOT, potentially after
the EDF has been loaded and manipulated. In that case, i.e. when using the
EPOCH-ANNOT command, the number of rows (epochs) in the .eannot
file must match the number of epochs that currently exist in the
in-memory representation of the EDF (i.e. which may be different from
the on-disk version).
A common use of an .eannot file could be to store manually-scored sleep stages:
wake
wake
N1
N1
wake
N1
N1
N2
N2
... (etc) ...
MASK command, such as:
MASK if=wake
RESTRUCTURE
wake epochs from analysis.
Luna XML files
Luna uses a version of the NSRR annotation format that is similar to
the NSRR XML format, but is designed to be completely interchangeable
with the .annot format. This is the format generated by the
WRITE-ANNOTS command when the xml option is
specified.
First, the preamble, with all information encapsulated in an Annotations parent node:
<?xml version="1.0" encoding="UTF-8" standalone="no"?>
<Annotations>
<SoftwareVersion>luna-v0.25</SoftwareVersion>
<StartTime> xxx </StartTime>
<Duration> xxx </Duration>
<DurationSeconds> xxx </DurationSeconds>
<EpochLength>30</EpochLength>
Second, the definition of each annotation class present in the Classes and Class tags:
<Classes>
<Class>
<Name>Annotation Class Name</Name>
<Description>Annotation Description</Description>
<Variable name="name1" type="num">Numeric variable label</Variable>
<Variable name="name2" type="num">Numeric variable label</Variable>
<Variable name="name3" type="num">Numeric variable label</Variable>
</Class>
</Classes>
Finally, each instance is listed in the next Instances and Instance tags:
<Instances>
<Instance>
<Class>Recording Start Time</Class>
<Name>Recording Start Time</Name>
<Start>0</Start>
<Duration>32820.0</Duration>
<Channel>Optional channel label(s)</Channel>
<Value var="name1">0.1</Value>
<Value var="name2">0.2</Value>
<Value var="name3">0.3</Value>
</Instance>
</Instances>
</Annotations>
NSRR XML files
Luna accepts XML annotation
files, as used by the National Sleep Research
Resource. These are based on Compumedics
Profusion files, as described
here.
Any files that end in a .xml or .XML extension are assumed to be
in this format, and Luna will attempt to read it as an annotation
file. Luna maps each ScoredEvent node to a single annotation
class.
--xml
To quickly view the Scored Events in a single NSRR XML file (sorted by clock time):
luna --xml my-annotations.xml
. . EpochLength 30
0 - 30 (30 secs) SleepStage Wake
30 - 60 (30 secs) SleepStage Wake
60 - 90 (30 secs) SleepStage Wake
90 - 120 (30 secs) SleepStage Wake
120 - 150 (30 secs) SleepStage Wake
150 - 180 (30 secs) SleepStage NREM1
180 - 210 (30 secs) SleepStage Wake
210 - 240 (30 secs) SleepStage Wake
... (etc) ...
--xml2
Alternatively, use --xml2 to report all entries in any XML, along
with the full, original XML document tree structure:
EDF+ Annotations
Using a test EDF+ file that contains some annotations
(one posted on Teunis van Beelen's website, which can be accessed here),
Luna will automatically extract the annotations (by convention, from channels named EDF Annotations) and include
these alongside any other annotations (e.g. from XML or .annot files). For this example EDF+ file:
luna ma0844az_1-1+.edf -o out.db -s ANNOTS
EDF Annotations channel is present, and is being extracted:
signals: 38 (of 38) selected in an EDF+ file:
EEG_FP1 | EEG_FP2 | EEG_F3 | EEG_F4 | EEG_C3 | EEG_C4 | EEG_P3 | EEG_P4
EEG_O1 | EEG_O2 | EEG_F7 | EEG_F8 | EEG_T3 | EEG_T4 | EEG_T5 | EEG_T6
EEG_FZ | EEG_CZ | EEG_PZ | EEG_E | EEG_PG1 | EEG_PG2 | EEG_A1 | EEG_A2
EEG_T1 | EEG_T2 | EEG_X1 | EEG_X2 | EEG_X3 | EEG_X4 | EEG_X5 | EEG_X6
EEG_X7 | DC01 | DC02 | DC03 | DC04 | EDF Annotations
extracting 'EDF Annotations' track
annotations:
edf_annot (x41)
By default, note that all annotations from an EDF+ file are assigned the
annotation class of edf_annot; the annotation instance ID is
assigned to whatever value is present in the EDF+ (annotation instance
IDs need not be unique). See above for a description of annotation
class versus instances.
Assigning class-level annotations from EDF+ explicitly
Exceptions to the rule can be added with the edf-annot-class special variable, to give a list of annotations that are their own classes: e.g.
luna ma0844az_1-1+.edf -o out.db edf-annot-class=HVT_START,HVT_END -s ANNOTS
The console now shows these explicitly listed:
annotations:
HVT_END (x1) | HVT_START (x1) | edf_annot (x39)
ANNOTS output, and can more easily be used as annotations in
masks, etc. Typically this might be done to pull out sleep stage annotations: e.g. to pull out stages from an EDF+
(assuming these are the labels) and save as a simple .annot file:
luna edfplus.edf edf-annot-class=N1,N2,N3,R,W,? \
-s ' WRITE-ANNOTS file=a.annot annot=N1,N2,N3,R,W,? '
skip-edf-annots=T as a special variable to ignore annotations from an EDF+
that have already been extracted to a separate file and attached (and linked via a sample-list to the original EDF+).It can be slow to scan an entire EDF+ to pull out annotations, and so performing these steps initially (i.e. to extract all desired annotations as an
.annot) is
the best workflow in Luna.
If the verbose special variable is set to true, then (surprisingly...) you'd see more verbose output
in the log, that lists some of these instance IDs too:
annotations:
[edf_annot] 41 instance(s) (from ma0844az_1-1+.edf)
36 instance IDs: A1+A2_OFF HVT_00:30 HVT_01:00 HVT_01:30 HVT_02:00 ...
The ANNOTS command can be used to tabulate these annotations.
luna ma0844az_1-1+.edf -o out.db -s ANNOTS
destrat out.db +ANNOTS -r ANNOT INST T
(nb. below, omitting some columns for clarity of output, and just listing the first three annotations:)
ANNOT INST START START_ELAPSED_HMS START_HMS
edf_annot PAT_IIB_EEG 0 00.00.00 10.18.42
edf_annot REC_START_IIB_CAL 0 00.00.00 10.18.42
edf_annot A1+A2_OFF 1 00.00.01 10.18.43
...
For each annotaton that start (and stop) times are given: in seconds elapsed since start of the EDF (START),
the same information but in hh:mm:ss format (START_ELAPSED_HMS) and as clock-time (START_HMS).
Spaces in annotation labels
In the original EDF+, the annotation labels are actually PAT IIB
EEG, REC START IIB CAL, etc. Note that Luna has autoamtically
converted spaces to underscore characters (_), to make working
with these annotations easier on the command line (i.e. PAT_IIB_EEG, etc). If for some reason this was
not desired, set the special variable keep-annot-spaces=T (or keep-spaces=T to do
this for both channel and annotation labels.
Luna masks typically refer to annotation class labels to work, but can also use annotation instance identifiers, which
will typically be necessary with EDF+ annotations (i.e. unless every annotation in the EDF meant the same thing, in which case the edf_annot
class label could be used).
For example, we see overall this EDF+ is just over 30 minutes in duration, so 60 30-second epochs:
Processing: ma0844az_1-1+.edf [ #1 ]
duration: 00.30.18 ( clocktime 10.18.42 - 10.49.00 )
We'll use two of the annotations in this file to extract only the
epochs containing those annotations: HVT_00:30 and HVT_01:00,
which occur at 00.21.32 and 00.22.02 (elapsed time) respectively,
as one can see from looking at the full output as the above ANNOTS
command, for example.
To mask epochs based on these two labels, we can use the following, using the
class[instance|instance]
|-delimited list, within square brackets [ and ] following the class ID).
luna ma0844az_1-1+.edf -o out.db -s ' MASK mask-ifnot=edf_annot[HVT_00:30|HVT_01:00] '
Using eval expressions instead
This is a digression, but to illustrate a difference way to use Luna masks in this instance, and to point to a common problem/solution. Instead of the above form, to mask epochs based on these two labels, we could also use the following:
luna ma0844az_1-1+.edf
-o out.db
-s ' MASK expr=" edf_annot =~ c( {HVT_00:30} , {HVT_01:00} ) " '
CMD #1: MASK
options: expr=" edf_annot =~ c( {HVT_00:30} , {HVT_01:00} ) " sig=*
set epochs, to default length 30, 60 epochs
set masking mode to 'force'
based on eval expression [edf_annot =~ c( {HVT_00:30} , {HVT_01:00} )]
2 true, 58 false and 0 invalid return values
2 epochs match; 2 newly masked, 0 unmasked, 58 unchanged
total of 58 of 60 retained
Expressing the above MASK command on the command line in this way, we used -s to write it out. To stop the (bash) shell from interpreting special characters that
can often occur (e.g. & to separate Luna commands) we
placed all text within single quotes ('). We also had to place the eval expression with double quotes, so that Luna could correctly parse the entire expression.
This means we could not also use single quotes easily to specify string literals on the command line (e.g. HVT_00:30). We therefore used Luna's alternate way of
specifying string literals in eval expressions, using braces { and } -- so we have:
edf_annot =~ c( {HVT_00:30} , {HVT_01:00} )
edf_annot =~ c( 'HVT_00:30' , 'HVT_01:00' )
-s and therefore already enclosed in single quotes),
we would be able to use this second (more readable) form. Neither quotes nor braces are needed in the simpler format first given above.
REMAP
Remap annotation labels after loading
This command replicates the functionality of the remap command, but can be applied after
a dataset is first attached. It also allows for some flexibility in the format of the remapping file.
Parameters
| Parameter | Example | Description |
|---|---|---|
file |
f1.txt |
Required file with mappings |
remap-col |
T or F |
Optionally, specify whether a remap column is present |
optional-remap-col |
Allows but does not require a remap column |
|
allow-spaces |
Optionally, allow space as well as tab delimiters | |
verbose |
Verbose output to the console |
Output
No direct output, except for messages to the console log if verbose is specified.
Any remappings will be available for output via the ALIASES command however.
Example
If f.txt is a tab-delimited file containing:
remap A|B|C
remap Y|Z
then the following command would map any annotations B or C to
A, and any annotations Z to Y:
luna s.lst -s ' REMAP file=f.txt remap-col=T & ANNOTS '
This is equivalent to including the file as follows:
luna s.lst @f.txt -s ' ANNOTS '
Alternatively, if f.txt was instead space-delimited and as follows:
A B|C
Y Z
luna s.lst -s ' REMAP file=f.txt remap-col=F allow-spaces & ANNOTS '
f.txt as:
A|B|C
Y|Z
A and Y will be the primary annotation labels used.
Standard rules apply with respect to spaces, capitalization and special characters (i.e. this is not case-sensitive by default, etc)
ANNOTS
Tabulate and summarize annotation information
Produces information about the number and total duration of annotations in an EDF, at the whole-file level and optionally per-epoch.
By default, ANNOTS will only show annotations that span at least one
unmasked epoch. The definition of whether or not an annotation
instance is masked or not can be varied. Depending on the context,
this can be useful to generate different types of summaries, e.g. the
number of respiratory events in REM versus NREM sleep.
Parameters
| Parameter | Example | Description |
|---|---|---|
epoch |
epoch |
Show epoch-level summaries |
show-masked |
show-masked |
Show masked annotations (default is not to do so) |
any |
any |
Keep annotations that have any overlap with one or more unmasked epochs (default) |
all |
all |
Only keep annotations that are completely within unmasked epochs |
start |
start |
Keep annotations that start in an unmasked epoch |
The epoch option lists, for each epoch, all the annotations
that overlap with that epoch, given the specified overlap definition
(any, all or start). If the show-mask option is given as well,
all epochs and annotations are shown; otherwise, only unmasked epochs
and annotations are shown. The output contains two variables
(EPOCH_MASK and ANN_MASK) that indicate whether a given annotation
instance is masked or not.
Output
Class-level annotation summary (strata: ANNOT)
| Variable | Description |
|---|---|
COUNT |
Number of instances of that annotation class |
DUR |
Combined duration (seconds) of all instances of that annotation class (does not account for potential overlap) |
Instance-level annotation summary (strata: ANNOT x INST)
| Variable | Description |
|---|---|
COUNT |
Number of instances of that annotation class and instance ID |
DUR |
Combined duration (seconds) of all instances of that annotation class and instance ID (does not account for potential overlap) |
Instance-level annotation tabulation (strata: ANNOT x INST x T)
| Variable | Depends on | Description |
|---|---|---|
START |
Start time (seconds) of this instance | |
STOP |
Stop time (seconds) of this instance | |
VAL |
The meta-data for this instance, if any exists (otherwise missing NA) |
|
ALL_MASKED |
show-masked |
|
ALL_UNMASKED |
show-masked |
|
SOME_MASKED |
show-masked |
|
SOME_UNMASKED |
show-masked |
|
START_MASKED |
show-masked |
Per-epoch instance-level annotation tabulation (strata: E x INTERVAL x INST)
| Variable | Depends on | Description |
|---|---|---|
ANNOT_MASK |
epoch |
Flag whether this annotation instance is included or excluded (1 means masked or excluded) |
EPOCH_MASK |
epoch |
Flag whether this epoch is included or excluded (1 means masked or excluded) |
MAKE-ANNOTS
Create new annotations on-the-fly
This command makes new annotations, typically based on pairwise comparisons of existing annotations, that are:
-
union of two annotations
-
intersection of two annotations
-
only annotations A that overlap annotation B
-
only annotations A that do not overlap annotation B
Also, there are special cases to
-
add annotations corresponding to epochs (and optionally, delimiting the boundaries of those epochs) (
epoch) -
simply add a numbered annotation corresponding to each epoch (
epoch-num) -
merge contiguous or overlapping annotations of a single class (
flatten) -
cut up longer annotations along the lines of epoch boundaries (
split)
Parameters
Primary parameters
| Parameter | Example | Description |
|---|---|---|
annot |
annot=N |
Name of the new annotation |
expr |
expr=A+B |
A pairwise expression of two existing annotations A and B in form A*B, A|B, A+B, A-B |
epoch |
epoch=E |
Add a flattened annotation for all epochs |
epoch-num |
epoch-num=E |
Add annotations of numbered epochs |
flatten |
flatten=A |
Merge contiguous or overlapping epochs ( i.e. A|A) |
split |
split=A |
Split longer annotations into parts no longer than the underlying epochs |
Specify annot and either expr, flatten, split; alternatively, specify either epoch or epoch-num
Pairwise expressions (arguments for expr)
| Expression | Description |
|---|---|
A*B |
New annotation is the intersection of A and B (AND) |
A|B |
New annotation is the union of A and B (OR) |
A+B |
New annotation is events in A that are overlapped by B |
A-B |
New annotation is events in A that are not overlapped by B |
Misc. options for epoch
| Expression | Description |
|---|---|
w |
One or more edge window sizes (seconds) |
collapse-edges |
Do not distinguish left and right edges |
edge |
Optional replacement for edge string for edge annotation label |
Examples
Pairwise operations
Staring with the four-column (headerless) annotation file a.annot:
A . 10 20
A . 30 40
B . 32 38
A . 50 60
B . 45 65
C as follows: the intersection of A and B:
MAKE-ANNOTS annot=C expr=A*B
class start stop
C 32.000 38.000
C 50.000 60.000
A or B:
MAKE-ANNOTS annot=C expr=A|B
class start stop
C 10.000 20.000
C 30.000 40.000
C 45.000 65.000
A that are overlapped by at least one B:
MAKE-ANNOTS annot=C expr=A+B
class start stop
C 30.000 40.000
C 50.000 60.000
A that are not overlapped by any B:
MAKE-ANNOTS annot=C expr=A-B
class start stop
C 10.000 20.000
Flattening and splitting epochs
If a.annot is the following:
A . 10 20
A . 20 30
A . 30 40
A . 35 45
A . 60 180
MAKE-ANNOTS annot=C flatten=A
class start stop
C 10.000 45.000
C 60.000 180.000
Conversely, if attaching annotation C (as above) and running split to generate S:
MAKE-ANNOTS annot=S flatten=C
class start stop
S 10.000 30.000
S 30.000 45.000
S 60.000 90.000
S 90.000 120.000
S 120.000 150.000
S 150.000 180.000
That is, given (default) 30-second epochs, the annotations C are
split at epoch boundaries to make separate events. Note that the
second epoch only spans half an epoch (30 - 45 seconds).
Epoch numbering
To add a new annotation for each epoch starting EP_:
luna s.lst -s ' MAKE-ANNOTS epoch-num=EP & WRITE-ANNOTS file=a.annot '
class instance channel start stop meta
EP_0001 . . 0.000 30.000 .
W . . 0.000 10020.000 .
SpO2_artifact . SpO2 17.300 56.400 .
EP_0002 . . 30.000 60.000 .
EP_0003 . . 60.000 90.000 .
EP_0004 . . 90.000 120.000 .
EP_0005 . . 120.000 150.000 .
...
Epoch annotations
To demonstrate using epoch: for this EDF, when run on a standard (gapless) EDF it will
give a single annotation that encompasses all epochs:
MAKE-ANNOTS epoch=E
class start stop
E 0.000 41220.000
E annotation would
still reflect the same as above (i.e. 41220, not 41225 seconds duration).
If the EDF had gaps, this would be reflected in multiple E annotations:
MASK epoch=1-10,100-200 & RE & MAKE-ANNOTS epoch=E
class start stop
E 0.000 300.000
E 2970.000 6000.000
Considering a second scenario with more complex epoch structure:
as a demonstation, we select the first 30 seconds of an EDF and then
set 7-second epochs with 2-second increment, followed by MAKE-ANNOTS epoch:
MASK epoch=1 & RE & EPOCH len=7 inc=2 & RE & MAKE-ANNOTS epoch=E
class start stop
E 0.000 29.000
verbose to the EPOCH command as described here;
alternatively, here we can use MAKE-ANNOTS epoch-num to make new annotations corresponding
to each epoch:
class start stop
E_0001 0.000 7.000
E_0002 2.000 9.000
E_0003 4.000 11.000
E_0004 6.000 13.000
E_0005 8.000 15.000
E_0006 10.000 17.000
E_0007 12.000 19.000
E_0008 14.000 21.000
E_0009 16.000 23.000
E_0010 18.000 25.000
E_0011 20.000 27.000
E_0012 22.000 29.000
Other options for epoch
As secondary options (designed for some niche use cases, but presented here for completeness)
MASK epoch=1-10,20-30 & RE & MAKE-ANNOTS epoch=E w=5
class start stop
edge_left 0.000 5.000
E 0.000 300.000
edge_right 295.000 300.000
edge_left 570.000 575.000
E 570.000 900.000
edge_right 895.000 900.000
edge annotations are added to demarcate the edges of each
epoch annotation - i.e. regions near the bounadaries of the currently included
analytic interval. The label edge can be changed with the edge option; left
and right edges can be collapsed to one class with collapse-edges.
WRITE-ANNOTS
Output annotations in full Luna-format
Luna assumes that annotation data may arrive in subtly different
formats: the generic .annot format tries to make some allowances for
this, by making it easier to convert to .annot, for example:
- using (clock-time or elapsed) hh:mm:ss versus elapsed second versus epoch encoding
- ellipses to indicate that start continues until the next point
- the class versus instance ID distinction
- optional headers and columns (e.g. for channels or meta-data)
- ability to include dates in various formats for longer recordings
- meta-data for events allowed in various formats
When writing .annot files, Luna adheres to a standard, full default:
- standard six-column format
- extended eaders absent
- explicit start and stop times for annotations in elapsed seconds
- ordered by time of occurrence
- multiple annotation combined into a single
.annotfile - can output only a subset of annotations (via the
annotoption)
Optionally, one can instead write variations on the standard .annot form:
-
use clock-times instead of elapsed seconds (with
hms) -
dates and clock-times (with
dhms) -
tabular meta-data (with
tab-meta)
Alternatively, one can write XML files (with xml) instead of .annot,
although we suggest you use .annot format as the most practical
default.
Parameters
| Parameter | Example | Description |
|---|---|---|
file |
a.annot |
File name - should end if .annot (unless XML) |
annot |
N1,N2,N3,R,W |
Only output these options (versus all), this accepts wildcard * |
hms |
Write in hh:mm:ss output rather than elapsed seconds | |
dhms |
Write in dd-mm-yyyy-hh:mm:ss output rather than elapsed seconds | |
tab-meta |
T |
Write meta-data in tabular format |
meta |
F |
Do or don't write meta column 6 (writes . if F) |
xml |
Write to XML annotation format instead of .annot |
Secondary parameters
| Parameter | Example | Description |
|---|---|---|
headers |
Include verbose header rows in .annot |
|
minimal |
Do not include main class... header row ( or min) |
|
specials |
Add special internal annotatons: e.g. epoch_len, etc |
|
collapse |
For a discontinuous EDF, collapse annotation times | |
add-ellipsis |
For zero-duration annotations, add ... as the second field, i.e. extend to the next annotation |
|
min-dur |
1 | Set a minimum duration of events (in seconds) for them to be output |
offset |
-0.22 | Apply an offset of, e.g. -0.22 seconds, to all annotations when writing out |
Output
A new annotation file, saved to disk, as either .annot or XML formats.
Example
Importantly, in the context of multi-sample processing, the ^
wildcard character (which substitutes the individual's ID) will
typically be necessary:
WRITE-ANNOTS file=annots/^.annot
will write annots/p01.annot, annots/p02.annot, etc, for samples
with IDs p01, p02, etc.
If there are other annotations (e.g. B1, B2, etc) but you only
want WRITE-ANNOTS to output a subset, you can write
WRITE-ANNOTS file=a.annot annot=A1,A2,A3
or equivalently in this case, using a wildcard to match all annotations starting with A:
WRITE-ANNOTS file=a.annot annot=A*
Note: wildcards can only come at the end of annotation names:
e.g. annot=A_*_X would not be parsed as a wildcard, whereas
annot=A_X_* would be.
Collapsing EDF+D annotations: If the current data are in EDF+D
*(with gaps), adding the collapse option will splice out those gaps,
as shown in this
vignette. Note that if
combined in the same Luna run as a WRITE command, you should place
WRITE-ANNOTS collapse first, as if the EDF+D isn't truly
discontinuous (i.e. it may skip initial epochs, but is still a single
contiguous interval), then WRITE will convert the record to a
standard EDF in memory (without changing the annotation times).
META
Add meta-data to annotations based on other annotations or signals
This flexible command can add meta-data tags to existing (in-memory) annotations based on either the properties of signals or other annotations.
It operates in one of two primary modes: appending new values based on 1) other annotations (e.g. overlap or distinance to other annotation classes) or 2) the EDF signals that span those annotations (e.g. the mean value of a window around each annotation). A further special mode can add meta-data based on the duration of annotations.
Added annotation meta-data can be exported (with WRITE-ANNOTS) or used to derive other summary metrics.
Parameters
Annotation mode options (other, i.e. meta-data reflect overlaps with other annotations):
| Parameter | Example | Description |
|---|---|---|
annot |
X |
add meta-data to annotation X |
other |
A,B,C |
one or more other annotations |
md |
D |
key name for metadata (i.e. the main output of META) |
w |
10 | optional window size around X |
w-left |
10 | optional window size before X |
w-right |
10 | optional window size after X |
flatten |
make union (& join contiguous) other annotations |
Annotation mode functions: one of these to be selected, determines values of meta-data:
| Parameter | Description |
|---|---|
overlap |
any overlap with 1+ other annotation (0/1) |
complete-overlap |
is X completely spanned by 1+ other annotation (0/1) |
whole-other |
completely other completely spanned by X (0/1) |
count |
count number of instances of other (N) |
nearest |
distance (time in sec) to nearest other (0 if overlap) |
nearest-midpoint |
as above, but based on annotation mid-points |
nearest-start |
as above, but based on annotation starts |
nearest-stop |
as above, but based on annotation stops |
Signal mode options: (sig, i.e. meta-data reflect summaries of spanned signals):
| Parameter | Example | Description |
|---|---|---|
annot |
X |
add meta-data to annotation X |
sig |
S1 |
a single signal |
md |
D |
key name for metadata |
w |
10 | optional window size around X |
w-left |
10 | optional window size before X |
w-right |
10 | optional window size after X |
Signal mode functions (one of these to be selected, determines values of meta-data):
| Parameter | Description |
|---|---|
mean |
mean of signal interval spanned by annotation X |
min |
min of signal interval spanned by annotation X |
max |
max of signal interval spanned by annotation X |
range |
range of signal interval spanned by annotation X |
Special duration mode (dur, i.e. meta reflects event duration)
| Parameter | Example | Description |
|---|---|---|
annot |
X |
Add meta-data to annotation X |
dur |
Sets special mode, to add event duration as the meta-data value | |
md |
D |
Use key name D for meta-data |
Output
No output is generated (other than appending meta-data tags to existing in-memory annotations).
Example
Consider these partial example annotation files (all belonging to the same recording):
head *annot
Some respiratory events in a.annot with meta-data:
==> a.annot <==
H. Obstructive 4 . 85171.386 85186.474 SpO2DeltaE=4;ArE=0
H. Obstructive 4 . 85338.220 85356.166 SpO2DeltaE=2;ArE=0
A. Obstructive 2 . 85393.791 85406.958 SpO2DeltaE=6;ArE=1
H. Obstructive 4 . 85621.544 85632.661 SpO2DeltaE=4;ArE=0
H. Obstructive 4 . 85803.149 85814.584 SpO2DeltaE=3;ArE=0
H. Obstructive 4 . 85934.409 85975.542 SpO2DeltaE=3;ArE=0
A. Obstructive 2 . 85981.168 85995.404 SpO2DeltaE=4;ArE=0
A. Obstructive 2 . 86138.694 86152.374 SpO2DeltaE=6;ArE=1
A. Obstructive 2 . 86668.490 86682.708 SpO2DeltaE=7;ArE=0
H. Obstructive 4 . 86761.592 86772.709 SpO2DeltaE=3;ArE=1
Some staging in stages.annot (note, using a different date/time format):
==> stages.annot <==
Wake . . 20-Jun-2023 23:00:30 20-Jun-2023 23:01:00 .
Wake . . 20-Jun-2023 23:01:00 20-Jun-2023 23:01:30 .
Wake . . 20-Jun-2023 23:01:30 20-Jun-2023 23:02:00 .
Wake . . 20-Jun-2023 23:02:00 20-Jun-2023 23:02:30 .
Wake . . 20-Jun-2023 23:02:30 20-Jun-2023 23:03:00 .
Wake . . 20-Jun-2023 23:03:00 20-Jun-2023 23:03:30 .
Wake . . 20-Jun-2023 23:03:30 20-Jun-2023 23:04:00 .
Wake . . 20-Jun-2023 23:04:00 20-Jun-2023 23:04:30 .
Wake . . 20-Jun-2023 23:04:30 20-Jun-2023 23:05:00 .
Wake . . 20-Jun-2023 23:05:00 20-Jun-2023 23:05:30 .
And some position information in pos.annot:
==> pos.annot <==
Supine . . 20-Jun-2023 23:00:05 20-Jun-2023 23:35:45 .
Left . . 20-Jun-2023 23:35:45 20-Jun-2023 23:35:50 .
Prone . . 20-Jun-2023 23:35:50 20-Jun-2023 23:35:55 .
Supine . . 20-Jun-2023 23:35:55 21-Jun-2023 00:06:35 .
Right . . 21-Jun-2023 00:06:35 21-Jun-2023 00:06:40 .
Upright . . 21-Jun-2023 00:06:40 21-Jun-2023 00:06:50 .
Supine . . 21-Jun-2023 00:06:50 21-Jun-2023 00:06:55 .
Left . . 21-Jun-2023 00:06:55 21-Jun-2023 00:07:00 .
Supine . . 21-Jun-2023 00:07:00 21-Jun-2023 00:07:30 .
Left . . 21-Jun-2023 00:07:30 21-Jun-2023 00:07:35 .
The respiratory event annotations include two meta-data tags:
SpO2DeltaE and ArE. At this point, it doesn't matter what these
are: the META command is generic. Consider we have a sample list
s.lst linking these annotation files:
id1 . a.annot,pos.annot,stages.annot
Note that in this particular example the EDF field happens to be blank
(.), meaning that Luna will create an empty EDF; although this
won't have any signal data, it allows working with annotation files,
although we'll need to specify some start-times and dates below. This
is just to illustrate that is is possible to apply Luna to
annotation-only style datasets.
Also consider we have a remapping file param to give new annotation labels, which is necessary
to work with them programmatically, as below.
remap apnea|A. Obstructive
remap hypopnea|H. Obstructive
apnea and hypopnea (i.e. omitting spaces).
Here we attach and merge the annotations (in this example case,
fixing a dummy EDF start time and length, etc); we then add the duration (tag DUR) of each event
as a meta-data tag, for apnea, hypopnea and N2 and export those to the file f.annot. We
also use DERIVE (described elsewhere) to calculate the sum of all newly added
apnea event durations, and save this in the individual-level variable ADUR, which can then be output
by DERIVE:
luna s.lst @param -o out.db \
--date=20-Jun-2023 --time=00:00:00 --nr=500000 --rs=100 \
-s ' META annot=apnea,hypopnea,N2 dur md=DUR
WRITE-ANNOTS file=f.annot
DERIVE var=ADUR expr=" ADUR = sum( apnea.DUR ) " '
Here are some rows of f.annot, that now have all events merged into a single file
and with the DUR tag added to the requested annotations:
N1 . . 85020.000 85050.000 .
N1 . . 85050.000 85080.000 .
N2 . . 85080.000 85110.000 DUR=30
N2 . . 85110.000 85140.000 DUR=30
N2 . . 85140.000 85170.000 DUR=30
N2 . . 85170.000 85200.000 DUR=30
hypopnea 4 . 85171.386 85186.474 ArE=0;DUR=15.088;SpO2DeltaE=4
N2 . . 85200.000 85230.000 DUR=30
N2 . . 85230.000 85260.000 DUR=30
N2 . . 85260.000 85290.000 DUR=30
N2 . . 85290.000 85320.000 DUR=30
N2 . . 85320.000 85350.000 DUR=30
hypopnea 4 . 85338.220 85356.166 ArE=0;DUR=17.946;SpO2DeltaE=2
N2 . . 85350.000 85380.000 DUR=30
N2 . . 85380.000 85410.000 DUR=30
apnea 2 . 85393.792 85406.959 ArE=1;DUR=13.167;SpO2DeltaE=6
N2 . . 85410.000 85440.000 DUR=30
The final ADUR value is just over 3600 seconds (i.e. ~one hour).
destrat out.db +DERIVE
ID ADUR
id1 3628.389
Note that if we added meta=F the meta-data would not be written as key=value pairs to the sixth field;
also, adding tab-meta adds .annot fields after the sixth, for these meta-data values, which can be
easier to parse in some circumstances:
class instance channel start stop meta ArE DUR SpO2DeltaE
...
N1 . . 84990.000 85020.000 . . . .
N1 . . 85020.000 85050.000 . . . .
N1 . . 85050.000 85080.000 . . . .
N2 . . 85080.000 85110.000 . . 30 .
N2 . . 85110.000 85140.000 . . 30 .
N2 . . 85140.000 85170.000 . . 30 .
N2 . . 85170.000 85200.000 . . 30 .
hypopnea 4 . 85171.386 85186.474 . 0 15.088 4
N2 . . 85200.000 85230.000 . . 30 .
N2 . . 85230.000 85260.000 . . 30 .
N2 . . 85260.000 85290.000 . . 30 .
N2 . . 85290.000 85320.000 . . 30 .
N2 . . 85320.000 85350.000 . . 30 .
hypopnea 4 . 85338.220 85356.166 . 0 17.946 2
N2 . . 85350.000 85380.000 . . 30 .
N2 . . 85380.000 85410.000 . . 30 .
apnea 2 . 85393.792 85406.959 . 1 13.167 6
N2 . . 85410.000 85440.000 . . 30 .
...
As a second example, we'll add meta-data to indicate whether respiratory
events are NREM/REM, and supine or not, using META in annotation-overlap mode:
luna s.lst @param \
--date=20-Jun-2023 --time=00:00:00 --nr=500000 --rs=100 \
-s ' META annot=apnea,hypopnea flatten complete-overlap other=N1,N2,N3 md=NR
META annot=apnea,hypopnea complete-overlap other=Supine md=Supine
WRITE-ANNOTS file=f.annot '
The first META statement specifies all NREM epochs and "flattens" them, i.e. makes
a single event for all contiguous (or overlapping) instances of the listed other annotations.
This means that when we ask for complete-overlap (i.e. that the respiratory event is completely
spanned by NREM, this will include cases that span, e.g. N2 and N3 epochs, as otherwise
the respiratory event would not be defined as being completely spanned by a single other event).
This sets the tag to NR which will have a 0 or 1 value for each respiratory event, depending on
whether it is NREM or not, based on this definition.
The second META statement does the same for supine events; here we
don't need to flatten annotations, as there is only a single class
(Supine) and these position annotations aren't epoched (i.e. split
into smaller periods, as the staging data are, i.e. as separate
30-second events).
Now if we look at rows of f.annot we see the new NR and Supine tags: e.g.
R . . 107310.000 107340.000 .
hypopnea 4 . 107320.156 107355.889 ArE=0;NR=0;SpO2DeltaE=9;Supine=1
R . . 107340.000 107370.000 .
hypopnea 4 . 107362.718 107394.640 ArE=0;NR=0;SpO2DeltaE=8;Supine=1
R . . 107370.000 107400.000 .
W . . 107400.000 107430.000 .
W . . 107430.000 107460.000 .
N1 . . 107460.000 107490.000 .
apnea 2 . 107462.050 107488.069 ArE=0;NR=1;SpO2DeltaE=11;Supine=1
N1 . . 107490.000 107520.000 .
apnea 2 . 107508.588 107537.016 ArE=1;NR=1;SpO2DeltaE=13;Supine=1
N1 . . 107520.000 107550.000 .
As a final more complex example of DERIVE: here we calculate the AHI
(count of apnea events per hour) for events that are a) in N2, b)
associated with an arousal (as in this particular case, an ArE of 1
indicates this, and c) are associated with a desaturation of at least
4% (here, the precalculated SpO2DeltaE of 4 or more):
luna s.lst @param -o out.db \
--date=20-Jun-2023 --time=00:00:00 --nr=500000 --rs=100 \
-s ' META annot=apnea,hypopnea,N2 dur md=DUR
META annot=apnea,hypopnea flatten complete-overlap other=N2 md=N2
META annot=apnea,hypopnea complete-overlap other=Supine md=Supine
DERIVE var=my_ahi expr=" D = sum(N2.DUR) / 3600.0 ;
N = length(apnea[apnea.N2==1 && apnea.ArE==1 && apnea.SpO2DeltaE>=4 ] ) ;
my_ahi = N / D " '
The META commands add the tags as above, that can be used in the subsequent DERIVE expression:
-
DUR- duration of each annotation -
NR- a flag (0/1) for whether the respiratory event was NREM -
Supine- a flag (0/1) for whether the event was during a supine position
The DERIVE expression has three parts:
-
D = sum(N2.DUR)/3600 ;creates a new variableDwhich is the total N2 duration in hours; note how these expressions can access the meta-data values of annotations uses theclass.metasyntax -
N = length(apnea[apnea.NR == 1 && apnea.ArE == 1 && apnea.SpO2DeltaE >= 4 ] ) ;counts the number of apnea events that are flagged as N2 (hereNR), have an arousal (hereArEmeta-tag of 1) andSpO2DeltaEtag of at least 4 -
my_ahi = N / Dcreate a final (scalar) variable which is the count per hour (i.e. AHI under this definition).
The final output my_ahi as requested by the option var=my_ahi to DERIVE can be accessed in the output:
destrat out.db +DERIVE
ID my_ahi
id1 2.70198675496689
Although this particular example uses an empty EDF with fixed start
times, etc, the META and DERIVE commands can be used in the usual
multi-sample project-based manner, i.e. these derived metrics could be
calculated across large numbers of studies with a single command.
One thing to watch out for when working with expressions and
annotation meta-data is the treatment of missing values - DERIVE in
principle detects and handles missing values, but the logic of the
expression may not be as expected under some conditions. See the page
on eval expressions for more details.
SPANNING
Summarize the coverage of an EDF by one or more annotations
Produces information about the coverage of an EDF by a group of one or more annotations. This can be used to check whether sleep stage annotations cover the whole recording or not; it will also report on annotations that are outside the duration of the EDF, and whether any of the annotations in the group overlap each other.
The report treats all annotations in the group as identical, asking what proportion of the EDF is spanned by one or more of the specified annotations.
Note
Currently, this command can only be applied to continuous,
unmasked EDFs (i.e. not discontinuous EDF+ files, not EDFs after
running the MASK or RE commands.
Parameters
| Parameter | Example | Description |
|---|---|---|
annot |
annot=N1,N2,N3,R,W |
Annotation(s) to group and report on |
Output
Class-level annotation summary (strata: none )
| Variable | Description |
|---|---|
REC_HMS |
EDF recording duration (hh:mm:ss) |
REC_SEC |
EDF recording duration (seconds) |
ANNOT_N |
Number of annotations in group |
ANNOT_SEC |
Total (potentially overlapping) annotation duration (secs) |
ANNOT_HMS |
Total (potentially overlapping) annotation duration (hh:mm:ss) |
ANNOT_OVERLAP |
Do any annotations in group overlap w/ one another (0/1)? |
VALID_N |
Number of valid annotations, ANNOT_N - INVALID_N |
INVALID_N |
Number of annotations that over-extend EDF duration |
INVALID_SEC |
Total duration of all annotation beyond EDF end |
SPANNED_SEC |
Duration of EDF spanned by 1+ of these annotations (secs) |
SPANNED_HMS |
Duration of EDF spanned by 1+ of these annotations (hh:mm:ss) |
SPANNED_PCT |
% of EDF spanned by 1+ of these annotations |
UNSPANNED_SEC |
Duration of EDF unspanned by 1+ of these annotations (secs) |
UNSPANNED_HMS |
Duration of EDF unspanned by 1+ of these annotations (hh:mm:ss) |
UNSPANNED_PCT |
% of EDF unspanned by 1+ of these annotations |
List of invalid annotations (strata: N )
| Variable | Description |
|---|---|
ANNOT |
Annotation class |
INST |
Annotation instance ID |
START |
Start (seconds) |
STOP |
Stop (seconds) |
Example
Here we use SPANNING on the first tutorial
individual. For example, given the sleep stage annotations, we might
want to check that all epochs are spanned by one (and only one) stage
annotation:
luna s.lst 1 -o out.db -s SPANNING annot=NREM1,NREM2,NREM3,REM,wake
destrat out.db +SPANNING | behead
ID nsrr01
ANNOT_HMS 11:21:30
ANNOT_N 1363
ANNOT_OVERLAP 0
ANNOT_SEC 40890
INVALID_N 0
INVALID_SEC 0
REC_HMS 11:22:00
REC_SEC 40920
SPANNED_HMS 11:21:30
SPANNED_PCT 99.9266862170088
SPANNED_SEC 40890
UNSPANNED_HMS 00:00:30
UNSPANNED_PCT 0.0733137829912023
UNSPANNED_SEC 30.0
VALID_N 1363
The above tells us that 99.93% (SPANNED_PCT) of the EDF is spanned
by one of these annotations. Furthermore, there is no overlap among
this set of annotations (ANNOT_OVERLAP is 0). See do we do see
there is 30 seconds (i.e. one epoch) not covered by any of these
annotations (UNSPANNED_SEC, i.e. 0.07% as noted by UNSPANNED_PCT).
Looking at the console/log output, we can see why this is the case: we omitted NREM4
which is also present as a staging annotation in this case:
[NREM1] 109 instance(s) (from edfs/learn-nsrr01-profusion.xml)
[NREM2] 523 instance(s) (from edfs/learn-nsrr01-profusion.xml)
[NREM3] 16 instance(s) (from edfs/learn-nsrr01-profusion.xml)
[NREM4] 1 instance(s) (from edfs/learn-nsrr01-profusion.xml)
[REM] 238 instance(s) (from edfs/learn-nsrr01-profusion.xml)
Re-running with this included now shows that these annoations a)
completely span the EDF (SPANNED_PCT is 100%) without any overlap
(ANNOT_OVERLAP is 0):
luna s.lst 1 -o out.db -s SPANNING annot=NREM1,NREM2,NREM3,NREM4,REM,wake
ID nsrr01
ANNOT_HMS 11:22:00
ANNOT_N 1364
ANNOT_OVERLAP 0
ANNOT_SEC 40920
INVALID_N 0
INVALID_SEC 0
REC_HMS 11:22:00
REC_SEC 40920
SPANNED_HMS 11:22:00
SPANNED_PCT 100
SPANNED_SEC 40920
UNSPANNED_HMS 00:00:00
UNSPANNED_PCT 0
UNSPANNED_SEC 0
VALID_N 1364
As an alternative (and to demonstrate using Luna in different ways), we
could have identified the "unspanned" epoch and its annotations as
follows, using the ANNOTS command (described above) on the
EDF after excluding epochs with one of the above five annotations:
luna s.lst 1 -o out.db -s 'MASK mask-if=NREM1,NREM2,NREM3,REM,wake & RE & ANNOTS'
NREM4 annotation:
destrat out.db +ANNOTS -r ANNOT
ID ANNOT COUNT DUR
nsrr01 NREM4 1 30
nsrr01 hypopnea 1 15.3
ESPAN
Epoch-based annotation coverage summaries
Produces information about the epoch-by-epoch coverage one or more annotations. This gives output for both individual annotation classes, as well as the group of annotations as a whole. It can output the following:
- number of seconds spanned by an annotation (per epoch)
- an indicator for whether any annotation spans that epoch
- the proportion of the epoch spanned by annotations
- the count of unqiue events spanning that epoch
Outputs are generated both per annotation class, and as a single, combined set (pooling all annotation classes).
Annotations are "flattened" (i.e. overlapping events merged) when calculating these statistics.
Parameters
| Parameter | Example | Description |
|---|---|---|
annot |
annot=N1,N2,N3,R,W |
Annotation(s) to report on |
sec |
F |
Report seconds of coverage (default: T) |
pct |
F |
Report percent of coverage (default: F) |
cnt |
F |
Report number of spanning annotations(default: F) |
has |
F |
Report presence/absence (0/1) (default: F) |
Output
Epoch-level summary (strata: E)
| Variable | Description |
|---|---|
SEC |
Total duration (seconds) (default, unless sec=F) |
PCT |
Proportion of epoch spanned by any of the listed annotations (if pct set) |
HAS |
Absence/presence (0/1) for any event spanning this epoch (if has set) |
CNT |
Count of any events spanning this epoch (if cnt set) |
Class-level annotation summary (strata: E x ANNOT )
| Variable | Description |
|---|---|
SEC |
Total duration (seconds) (default, unless sec=F) |
PCT |
Proportion of epoch spanned by this annotation class (if pct set) |
HAS |
Absence/presence (0/1) for any event spanning this epoch (if has set) |
CNT |
Count of events spanning this epoch (if cnt set) |
Event-level annotation output (option: verbose; strata: E x ANNOT x INST )
| Variable | Description |
|---|---|
DUR |
Event duration |
START |
Event start (seconds) |
STOP |
Event stop (seconds) |
XDUR |
Duration of event within this epoch |
XSTART |
Start of event within this epoch |
XTOP |
Stop of event within this epoch |
Example
luna s.lst -o out.db -s ESPAN annot=hypopnea,arousal,desat
destrat out.db +ESPAN -r E -c ANNOT
ID E SEC.ANNOT_arousal SEC.ANNOT_desat SEC.ANNOT_hypopnea
...
id0 650 0 0 0
id0 651 0 0 0
id0 652 0 0 0
id0 653 0 0 0.2
id0 654 0 28 12.5
id0 655 6.4 22 14.5
id0 656 0 30.0 2.5
id0 657 0 2 0
id0 658 0 0 0
id0 659 0 0 0
id0 660 0 0 0
id0 661 0 0 0
...
Re-running with additional options to a) give the three other outputs, and b) output event-instance information:
luna s.lst -o out.db \
-s ESPAN annot=hypopnea,arousal,desat pct sec cnt has verbose
Here showing epoch/annotation level output (just for a few epochs, and
here only for hypopnea):
destrat out.db +ESPAN -r E ANNOT
ID ANNOT E INST DUR START STOP XDUR XSTART XSTOP
...
id0 hypopnea 653 1 12.7 19589.8 19602.5 0.2 19589.8 19590
id0 hypopnea 654 1 12.7 19589.8 19602.5 12.5 19590 19602.5
id0 hypopnea 655 1 17 19635.5 19652.5 14.5 19635.5 19650
id0 hypopnea 656 1 17 19635.5 19652.5 2.5 19650 19652.5
...
Here is some epoch-level output, combined across all three annotation classes:
destrat out.db +ESPAN -r E
ID E CNT HAS PCT SEC
...
id0 650 0 0 0 0
id0 651 0 0 0 0
id0 652 0 0 0 0
id0 653 1 1 0.0066 0.2
id0 654 2 1 1 30.0
id0 655 4 1 0.96 28.8
id0 656 2 1 1 30.0
id0 657 1 1 0.0666 2
id0 658 0 0 0 0
id0 659 0 0 0 0
id0 660 0 0 0 0
id0 661 0 0 0 0
...
The four events that span epoch 655 (CNT above) can be obtained from
the E x ANNOT x INST event-level output: e.g.
ID ANNOT E INST DUR START STOP XDUR XSTART XSTOP
id0 arousal 655 1 6.4 19627.9 19634.3 6.4 19627.9 19634.3
id0 hypopnea 655 1 17 19635.5 19652.5 14.5 19635.5 19650
id0 desat 655 1 38 19592 19630 10 19620 19630
id0 desat 655 2 44 19638 19682 12 19638 19650
Combining EPOCH and ESPAN
It can often be helpful to also
run EPOCH verbose (or, if epochs are already defined and you
just want to list them, not change/set them, EPOCH table) prior
to ESPAN: this will dump the start/stop times of the epochs,
that can be linked to the output of ESPAN via the shared E
field per individual.
Looking at the outputs of this will often correspond to how annotation-based mask commands would work. As a contrived example, if we wanted to select only epochs with all three annotations:
luna s.lst -o out.db \
-s 'ESPAN annot=hypopnea,arousal,desat has=T sec=F'
ID E HAS.ANNOT_arousal HAS.ANNOT_desat HAS.ANNOT_hypopnea
...
id0 650 0 0 0
id0 651 0 0 0
id0 652 0 0 0
id0 653 0 0 1
id0 654 0 1 1
id0 655 1 1 1
id0 656 0 1 1
id0 657 0 1 0
id0 658 0 0 0
id0 659 0 0 0
id0 660 0 0 0
...
Separately running the implied mask command and looking at the epoch information afterwards:
luna s.lst -o out.db \
-s ' MASK ifnot-all=hypopnea,arousal,desat & RE & EPOCH table '
CMD #1: MASK
options: ifnot-all=hypopnea,arousal,desat sig=*
set epochs, to default length 30, 1346 epochs
set masking mode to 'force'
annots: arousal desat hypopnea
applied annotation mask for 3 annotation(s) (using and-matching across multiple annotations)
1 epochs match; 1345 newly masked, 0 unmasked, 1 unchanged
total of 1 of 1346 retained
Pulling the epoch information (not showing all columns below) we see that only
epoch 655 is retained, consistent with it being the only epoch from ESPAN to have (HAS==1)
for all three annotation classes:
destrat out.db +EPOCH -r E
ID E DUR E1 HMS START STOP
id0 655 30 1 02:13:03.000 19620 19650
A2S
Add a signal based on an annotation
The A2S command, by default, makes a binary (0/1) EDF channel
corresponding to one or more annotation(s), i.e. whether that
sample-point is spanned by that annotation or not. Alternatively, it
can use the instance ID if it is a numeric value (instead of always
1).
| Option | Example | Description |
|---|---|---|
annot |
arousal |
Name of annotation to select |
sr |
100 |
Sample rate of new signal |
label |
A |
Optionally, a name for the signal (otherwise is set to annot value) |
numeric-inst |
Sets signal to numeric value of the annotation instance IDs , not just 0/1 |
S2A
Add an annotation based on a signal
The complement to A2S, the S2A command makes an annotation
corresponding to ranged intervals in an EDF channel. For example,
sometimes body position is encoded as a signal with values 0,
1, 2 or 3, e.g. for supine, non-supine, left, right. The S2A
command can make an annotation that represents the same information
(e.g. and be saved with WRITE-ANNOTS or used in a MASK, etc.)
Parameters
| Option | Example | Description |
|---|---|---|
sig |
C3 |
Name of signal |
encoding |
S,0,NS,1,L,2,R,3 |
Signal encoding (see below) |
encoding2 |
X,5,10,Y,20,30 |
Alternate encoding (see below ) |
class |
position |
Make one annotation class (e.g. position) w/ labels as instance IDs |
span-gaps |
How to handle signal discontinuities |
Output
A new annotation is added to the internal store for this individual; otherwise, no explicit output is generated except for some notes to the log/console.
Example
In the NSRR CFS dataset, body
position is encoded as an EDF channel called POSITION. In
contrast, some other NSRR studies encode body position as an
annotation separate from the EDF.
First, to confirm the contents of this channel, we might extract POSITION
to a text file for one individual:
luna cfs.lst 1 -s MATRIX min file=pos.txt sig=POSITION
Plotting the values in pos.txt, we see that this signal contains
only 4 discrete values, here plotted across the night:
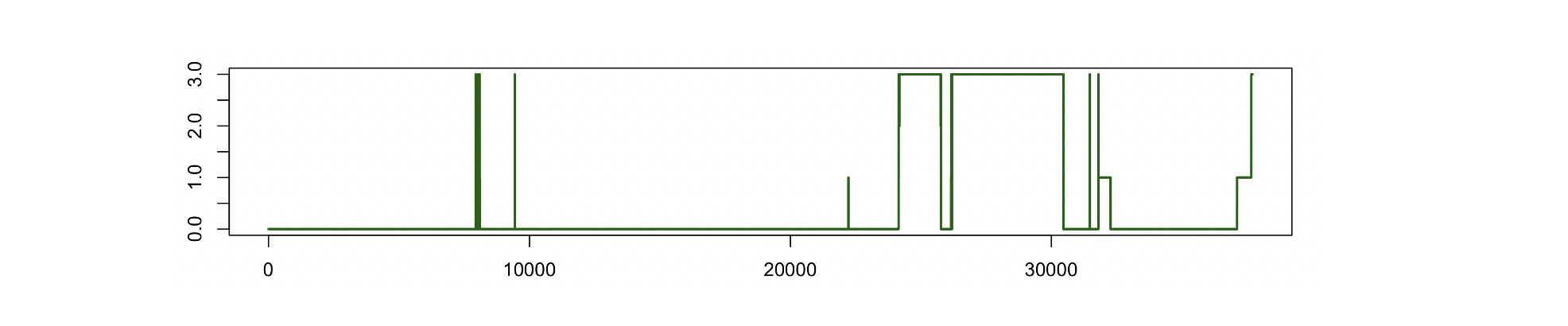
as tabulated here:
cat pos.txt | sort | uniq -c
30655 0
1059 1
66 2
5930 3
(i.e. the uniq -c command counts 30,655 instances of 0, 1,059
instances of 1, etc).
Let's say the CFS encoded body positions supine, non-supine, left and
right as 0, 1, 2 and 3 respectively. To generate annotations based on this signal,
we can use the S2A command:
luna cfs.lst -s ' S2A sig=POSITION encoding=S,0,NS,1,L,2,R,3
WRITE-ANNOTS file=pos-^.annot '
The generated .annot file(s) (note: using ^ swaps in the
individual's ID to make a filename unique to that individual) might
look like:
# L
# NS
# R
# S
class instance channel start stop meta
S . POSITION 0.000 7939.000 .
R . POSITION 7939.000 7940.000 .
S . POSITION 7940.000 7941.000 .
R . POSITION 7941.000 7943.000 .
... cont'd ...
That is: four new annotations (S, NS, L and R) have been generated from the values of the POSITION channel, as specified by
the encoding argument of S2A, which takes one or more label/value pairs as above (i.e. S=0, NS=1, etc).
Note that EDF is a floating-point numeric format, and depending on the
encoding of the EDF (physical and digital min/max in the headers), or
whether the channel has been processed or copied, etc, then the values
0, 1, 2 and 3 might not be numerically exactly 0.0, 1.0, etc
(i.e. given finite, 16-bit resolution of EDFs, where it typically is
irrelevant whether an EEG signal amplitude is, say, 15.08 microvolts
versus 15.07995 microvolts, as this level of numerical difference is
below the measurement accuracy of the original data).
In this type of scenario, where categorical (integer) information is
being represented by an inherently floating-point format, these
numerical differences might matter, however. Because of this, for a
value such as 1, the default encoding actually matches on a window
of 1.00 plus or minus 0.05 (i.e. 0.95 to 1.05). This is clear
in the output sent to the log file:
CMD #1: S2A
options: encoding=S,0,NS,1,L,2,R,3 sig=POSITION
encoding 4 annotation instances
added 9 intervals for L based on 1.95 <= POSITION <= 2.05
added 19 intervals for NS based on 0.95 <= POSITION <= 1.05
added 26 intervals for R based on 2.95 <= POSITION <= 3.05
added 37 intervals for S based on -0.05 <= POSITION <= 0.05
If for some reason this is not appropriate, or if you wish to match
larger intervals to a given annotation, then you can use the alternate
encoding specification, via the encoding2 option (instead of
encoding). The encoding2 argument takes two forms; the first takes
triplets of {label, value, window}:
encoding2=S,0,+0.05,NS,1,+0.05,L,2,+0.05,R,3,+0.05’
encoding
above; the point of this form is that you can specify values other
than 0.05 as the window size (note the use of + is necessary to
invoke this mid-point, window width encoding, i.e. +0.05 and not
just 0.05).
The second form of encoding2 takes a lower and upper bound (rather
than a mid-point and window width). For example, to give a different
example:
encoding2=X,5,10,Y,20,30’
which implies X maps to values between 5.0 and 10.0, whereas Y
maps to values between 20.0 and 30.0. Note that not all values have
to be covered (i.e. those points, such as a value of 15,
would have neither an X nor Y annotation assigned to span that point).
Note that the above form would of course be
identical to:
encoding2=X,7.5,+2.5,Y,15,+5
In the first example, S2A will by default add four new annotation
classes: S, NS, L and R corresponding to the labels in the
encoding. As noted in the example above, the instance IDs
will be blank (.). If you instead added:
class=pos
pos annotation would be added, and the instance
IDs would encode the type of position. i.e. the above example would
become:
# pos
class instance channel start stop meta
pos S POSITION 0.000 7939.000 .
pos R POSITION 7939.000 7940.000 .
pos S POSITION 7940.000 7941.000 .
pos R POSITION 7941.000 7943.000 .
...
Finally, by default if an EDF contains gaps (i.e. if it is an EDF+D,
or if it has been internally restructured via MASK/RE commands, etc)
then any new annotations will also stop/restart at those gaps. If the
span-gaps option is given, then the generated annotations will not
stop/restart at gaps, but will (guess what...) span those gaps
instead. For example, consider a dummy signal S with these time
points (T) and a gap (discontinuity) between time-points 4 and 8
(i.e. no valid values for S):
T = 1 2 3 4 5 6 7 8 9
S = 1 1 3 3 . . . 3 3
If 1 maps to NS and 3 maps to R as above, then by default the annotations generated would be:
class start stop
NS 1 2
R 3 4
R 8 9
span-gaps addded, then we would see:
class start stop
NS 1 2
R 3 9