Spindles and slow oscillations (SO)
| Command | Description |
|---|---|
SPINDLES |
Wavelet-based spindle detection |
SO |
Slow oscillation detection |
SPINDLES
Wavelet-based sleep spindle detection
This command detects spindles using a wavelet-based approach.
Optionally, it also detects slow oscillations (SO) and the temporal
coupling between spindles and SO. A target frequency is specified,
which corresponds to the center frequency of the Morlet wavelet. A
single SPINDLES command can detect spindles at different
frequencies, and on different channels; there are also options for
collating putative spindles that are coincident in time but
observed on different channels and/or at neighbouring frequencies.
The SPINDLES command has quite a large number of options (although
most of them are not necessary for its basic operation). For clarity,
this section is split into a number of sub-sections that describe
different aspects of this command:
- Basic usage
- Determining thresholds
- Collating spindles
- Additional spindle morphology metrics
- Generating spindle annotations and viewing spindles
- Spindle/SO coupling
After running the wavelet transform, spindles are detected based on the following rules:
-
the magnitude of the wavelet transform is smoothed (default sliding window of 0.1,
win) and scaled by the whole-signal mean (or median, if themedianflag is used) -
here, whole-signal means all unmasked epochs available to this command (i.e. may be only all N2 epochs rather than all epochs)
-
a spindle is called when a core interval of at least 0.3 seconds (
min0) exhibits a signal at least 4.5 times (th) the mean (or median) -
furthermore, all spindles must also exhibit an extended interval of at least 0.5 seconds (
min) with a signal at least 2 times (th2) the mean (or median) -
putative spindle intervals cannot be longer than 3 seconds (
max); further, spindles within 0.5 seconds (merge) are merged (unless the resulting spindle is greater thanmaxseconds) -
putative spindles are also filtered based on a quality metric q, the threshold of which is given by the
qparameter
Following this heuristic procedure, in addition to the count and density (count per minute) of spindles, the following quantities are estimated to characterize the morphology of each spindle, after bandpass-filtering the signal for FC ± 2 Hz:
-
spindle duration (of core plus flanking interval) in seconds, and number of oscillations
-
spindle amplitude, based on the maximum peak-to-peak amplitude
-
mean and maximum of the transformed wavelet statistic
-
spindle frequency, based on counting zero-crossings, and secondarily based on the modal frequency from a FFT
-
spindle symmetry, based on the relative location of the spindle's central "peak" (the point of maximum peak-to-peak amplitude), and a second "folded" symmetry index, calculated as 2|S-0.5|
-
spindle chirp, based on the log ratio of mean peak-to-peak time intervals in the first versus the second half of the spindle
-
a measure of integrated spindle activity (ISA), based on the sum of normalized wavelet coefficients in the spindle interval
-
a quality measure q based on the ratio of 1) the relative enrichment of power in the sigma band (for the spindle compared to baseline) versus 2) the comparable relative enrichment in power across all bands (i.e. again for the spindle compared to baseline)
Averaging over all spindles in a given run, Luna reports the mean spindle density, amplitude, duration, frequency, number of oscillations, chirp, symmetry indices, etc. Luna also reports per-spindle statistics, as well as per-epoch counts of spindles.
Basic usage
Parameters
The most basic parameter is fc, which specifies the target frequency
(or frequencies) for the wavelet(s). Combined with the cycles
parameter, this defines the distribution of spindle frequencies that
are targeted by the wavelet. See also the
CWT-DESIGN command, which gives further information on
wavelet properties.
The primary output variables for an individual are spindle density
(DENS), mean duration (DUR) and frequency (FRQ or FFT, both
are typically very similar). An additional metric of interest is
integrated spindle activity (ISA), which, for a given spindle, is
the sum of the normalized wavelet coefficients in the spindle
interval. Higher values indicate more evidence in favour of a
spindle, and longer and higher amplitude spindles. Averaging across
spindles detected, for an individual, ISAS is the
mean ISA, which reflects the average amplitude and duration of that
individual's spindles; ISAM is the mean ISA per
minute, which reflects the combination of spindle amplitude, duration
and density; ISAT is the total ISA, which
reflects the combination of spindle amplitude, duration and count.
Because it is normalized by the individual's baseline,
ISAS may be a better metric on which to base
inter-individual comparisons, compared to AMP which is more likely
to reflect technical differences in recordings.
Primary options
| Parameter | Example | Description |
|---|---|---|
sig |
sig=C3,F3 |
Restrict analysis to these channels (otherwise, all channels are included) |
fc |
fc=11,15 |
FC value(s), e.g. to find 11 and 15 Hz spindles (default 13.5 Hz) |
cycles |
cycles=12 |
Number of cycles, where more cycles gives better frequency but poorer temporal resolution (default 7) |
epoch |
epoch |
Show epoch-level output (number of spindles per epoch |
per-spindle |
per-spindle |
Show spindle-level output |
th |
th=6 |
Multiplicative threshold for core spindle detection (default 4.5) |
th2 |
th2=3 |
Multiplicative threshold for non-core spindle detection (default=2) |
median |
median |
Flag to indicate that the median, not mean, is used for thresholding |
q |
q=0.3 |
Quality metric criterion for individual spindles (default 0) |
Secondary parameters
Most users will not need to alter these.
| Parameter | Example | Description |
|---|---|---|
fc-lower |
fc-lower=9 |
Lower limit if iterating over multiple FC values |
fc-upper |
fc-upper=15 |
Upper limit if iterating over multiple FC values |
fc-step |
fc-step=2 |
Increment step if iterating over multiple FC values |
th-max |
th-max=10 |
Maximum threshold for spindle core (default: none) |
min |
min=1 |
Minimum duration for an entire spindle (default 0.5 seconds) |
min0 |
min0=0.3 |
Minimum duration for a spindle core (default 0.3 seconds) |
max |
max=2 |
Maximum duration for an entire spindle (default 3 seconds) |
win |
win=0.2 |
Smoothing window for wavelet coefficients (default 0.1 seconds) |
local |
local=120 |
Use local window (in seconds) to define baseline for spindle detection |
Cache options
| Parameter | Example | Description |
|---|---|---|
cache |
cache=w1 |
Cache CWT coefficients per sample point (e.g. for TLOCK) |
cache-peaks |
cache-peaks=p1 |
Cache spindle peaks (sample points) (e.g. for TLOCK) |
Characterizing external spindles
If you have an annotation based on spindles detected by an external method, you can
still use Luna to generate the same metrics for those events (i.e. rather than using its
own internal algorithm to detect those spindles), via the precomputed option. This naturally requires
that the events/spindles have been read in as an interval annotation.
| Parameter | Example | Description |
|---|---|---|
precomputed |
sp1 |
Use annotation class sp1 to define spindles, rather than have SPINDLES detect them |
Outputs
Individual-level output (strata: F x CH)
| Variable | Description |
|---|---|
DENS |
Spindle density (count per minute) |
N |
Total number of spindles |
AMP |
Mean spindle amplitude (uV or mV units) |
DUR |
Mean spindle duration (core+flanking region) |
NOSC |
Mean number of oscillations per spindle |
FWHM |
Mean spindle FWHM (full width at half maximum) |
ISA_S |
Mean integrated spindle activity (ISA) per spindle |
ISA_M |
Mean integrated spindle activity (ISA) per minute |
ISA_T |
Total integrated spindle activity (ISA) |
FRQ |
Mean spindle frequency (from counting zero-crossings) |
FFT |
Mean spindle frequency (from FFT) |
CHIRP |
Mean chirp metric per spindle |
SYMM |
Mean spindle symmetry metric |
SYMM2 |
Mean spindle folded-symmetry metric |
Q |
Mean spindle quality metric |
DISPERSION |
Mean dispersion index of epoch spindle count |
DISPERSION_P |
P-value for test of over-dispersion |
MINS |
Total duration of signal entered into the analysis (minutes) |
NE |
Number of epochs |
N01 |
Number of spindles prior to merging |
N02 |
Number of spindles post merging, prior to QC |
Epoch-level output (option: epoch; strata: E x F x CH)
| Variable | Description |
|---|---|
N |
Number of spindles observed in that epoch (for that target frequency/channel) |
Spindle-level output (option per-spindle; strata: SPINDLE x F x CH)
| Variable | Description |
|---|---|
AMP |
Spindle amplitude (uV or mV units) |
CHIRP |
Spindle chirp (-1 to +1) |
DUR |
Spindle duration (seconds) |
FWHM |
Spindle FWHM (seconds) |
NOSC |
Number of oscillations |
FRQ |
Spindle frequency based on counting zero-crossings in bandpass filtered signal |
FFT |
Spindle frequency based on FFT |
ISA |
Integrated spindle activity |
MAXTSTAT |
Maximum wavelet statistic |
MEANSTAT |
Mean wavelet statistic |
Q |
Quality metric |
PASS |
Flag (0/1) for whether this spindle passes the quality metric criterion |
START |
Start position of the spindle (seconds elapsed since start of EDF) |
STOP |
Stop position of the spindle (seconds elapsed since start of EDF) |
START_SP |
Start position of the spindle (in sample-units relative to current in-memory EDF) |
STOP_SP |
Stop position of the spindle (in sample-units relative to the current in-memory EDF) |
SYMM |
Symmetry index (relative position of peak) |
SYMM2 |
Folded symmetry index (0=symmetrical, 1=asymmetrical) |
Example
Here we estimate spindles for all NREM2 sleep for the
tutorial individual nsrr02:
luna s.lst nsrr02 -o out.db -s "MASK ifnot=NREM2 & RE & SPINDLES fc=11,15 sig=EEG"
We see some output is sent to the console describing the process. For slow (i.e. target frequency of 11 Hz) spindles:
CMD #3: SPINDLES
detecting spindles around F_C 11Hz
wavelet with 7 cycles
smoothing window = 0.1s
detection thresholds (core, flank, max) = 4.5, 2x
core duration threshold (core, min, max) = 0.3, 0.5, 3s
pruned spindle count from 296 to 275
filtering at 9 to 13
filtering channel EEG_BP_11_4, bandpass FIR order 71
estimated spindle density is 1.37845
The relevant strata in out.db are given by destrat, namely
per-individual, per-epoch and per-spindle respectively:
destrat out.db
[SPINDLES] : F CH : 2 level(s) : AMP CHIRP DENS DISPERSION DISPERSION_P
: : : DUR FFT FRQ FWHM ISA_M ISA_S ISA_T
: : : MINS N N01 N02 NE NOSC Q SYMM SYMM2
: : :
: : :
[SPINDLES] : E F CH : (...) : N
: : :
[SPINDLES] : F CH SPINDLE : 692 level(s) : AMP CHIRP DUR FFT FRQ FWHM ISA
: : : MAXSTAT MEANSTAT NOSC PASS Q START
: : : START_SP STOP STOP_SP SYMM SYMM2
: : :
To get the spindle density and mean spindle duration and frequency for this individual:
destrat out.db +SPINDLES -r F CH -v DENS DUR FFT -p 3
ID CH F DENS DUR FFT
nsrr02 EEG 11 1.378 0.847 11.280
nsrr02 EEG 15 2.090 0.847 13.475
We will further query the output using lunaR and
ldb(). In R:
library(luna)
k <- ldb( "out.db" )
We can use lx() to see the strata (factor combinations) that
correspond to the destrat output above:
lx(k)
MASK : EPOCH_MASK
RE : BL
SPINDLES : CH_F CH_E_F CH_F_SPINDLE
Looking at the per-epoch data:
d <- lx( k , "SPINDLES" , "CH_E_F" )
head(d)
> head(d)
ID CH E F N
1 nsrr02 EEG 92 11 2
2 nsrr02 EEG 92 15 0
3 nsrr02 EEG 93 11 4
4 nsrr02 EEG 93 15 0
5 nsrr02 EEG 98 11 2
6 nsrr02 EEG 98 15 1
Not that it is particularly informative in itself, but we can plot the number of spindles per epoch for fast and slow spindles:
par(mfcol=c(2,1), mar=c(4,4,1,1))
plot( d$E[ d$F == 11 ] , d$N[ d$F == 11 ] , pch= 20 , col="orange" ,
xlab="Epoch" , ylab="N spindles" , main="11 Hz spindles" )
plot( d$E[ d$F == 15 ] , d$N[ d$F == 15 ] , pch= 20 , col="skyblue" ,
xlab="Epoch" , ylab="N spindles" , main="15 Hz spindles" )
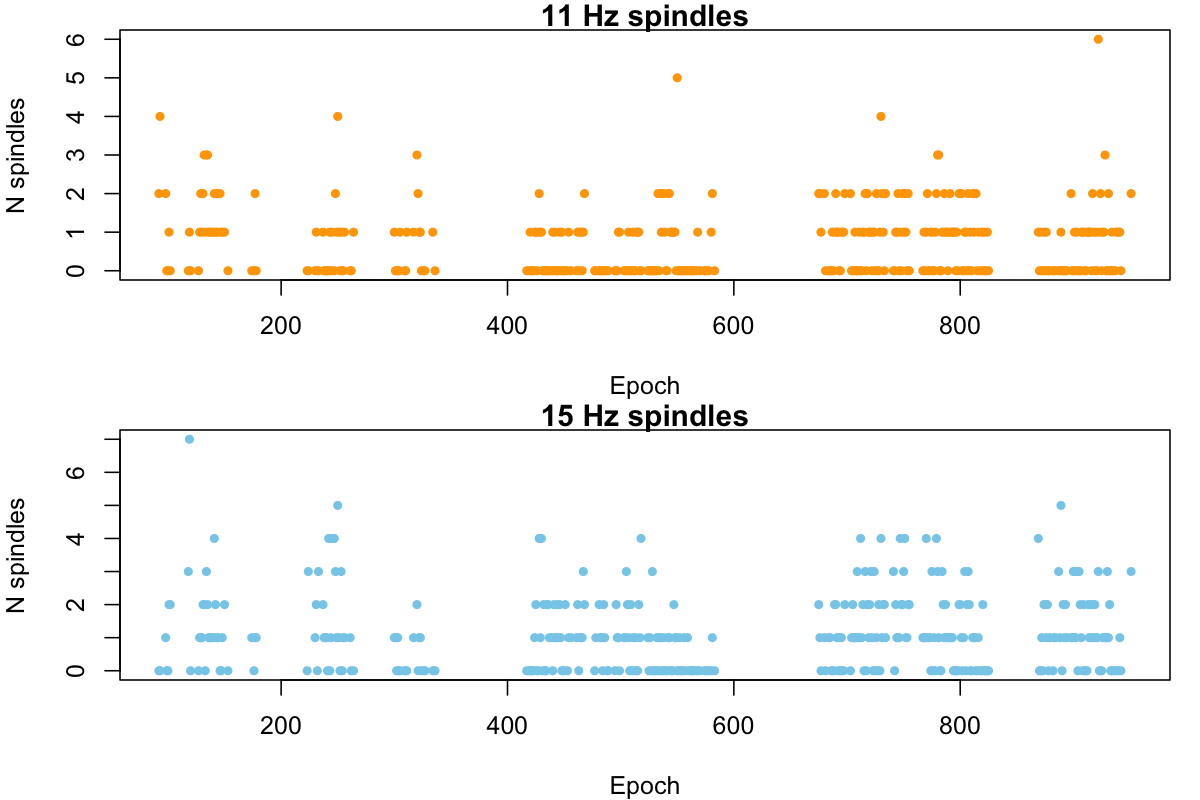
Luna provides a rough sanity-check to test for unusual distributions
of spindles per epoch, as might occur if a movement artifact, for
example, causes multiple spindles to be scored in one epoch. To a
first approximation, we can treat the distribution of spindles per
epoch as Poisson-distributed, and test for over-dispersion (i.e. if the variance
is significantly greater than the mean). This is provided by the DISPERSION and DISPERSION_P
variables in the individual-level output:
destrat out.db +SPINDLES -r F CH -v DISPERSION DISPERSION_P -p 3
ID CH F DISPERSION DISPERSION_P
nsrr02 EEG 11 1.143 0.606
nsrr02 EEG 15 1.243 0.038
In practice, we take values of DISPERSION above 2.0 to reflect
potentially problematic recordings/patterns of spindle detection, so these
don't look too bad. Potentially the epochs with more than five spindles may
questionable (especially as we did not apply any type of artifact correction prior
to detecting spindles):
table( d$F , d$N )
0 1 2 3 4 5 6 7
11 206 131 50 7 3 1 1 0
15 154 139 62 27 14 2 0 1
Finally, here is an example spindle detected in this individual, shown alongside the bandpass filtered EEG for delta, theta, alpha and beta power as well as sigma power. As expected (i.e. by definition), the spindle corresponds to a brief increase in sigma power:
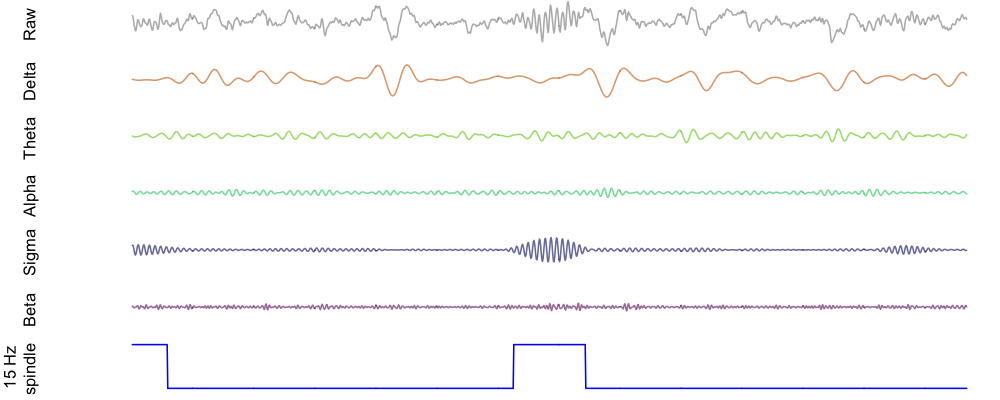
Note
Primarily in the context of lunaR, in the near future we'll be adding tutorials and helper functions to facilitate automatically making these types of plots quickly for all spindles in a dataset.
Thresholds
The core of the spindle detection algorithm involves imposing discrete thresholds on the quantitative metrics derived from the continuously-varying EEG. As such, there are a lot of magic numbers inherent in the spindle detection process. One of the key parameters is the threshold which the normalized wavelet coefficient must exceed for a putative spindle to be scored (the default of which is 4.5 times the mean). But is 4.5 a good magic number? What about 2 instead? Or what about 20? And so on...
To get some insight into whether this may be an appropriate choice for the particular individuals and analyses under consideration, Luna has a set of functions that evaluate different thresholds based on Otsu's method, a technique from computer vision that attempts to set a threshold so as to minimize the within-class variance (and so correspondingly, maximize the between class variance) in the two classes that are formed by placing a dichotomizing threshold on a continuous measure. In our scenario, the measure is the wavelet coefficient, and we are trying to place a threshold that roughly corresponds to "spindle" and "non-spindle".
If the continuous metric exhibits clear bimodality, this approach should easily find a threshold that effectively splits the two peaks. In the context of spindles, where things are likely to be a bit messier, we consider the distribution of wavelet coefficients to be a mixture of two classes: the null distribution of noise, i.e. where there is no true spindle, and a second, less frequency class representing points where a spindle is present (and where we will expect higher values of the wavelet coefficient).
Parameters
| Parameter | Example | Description |
|---|---|---|
empirical |
empirical |
Empirically determine thresholds |
set-empirical |
set-empirical |
Use empirically determined thresholds for spindle detection |
verbose-empirical |
verbose-empirical |
Output extensive information on threshold estimation |
Output
Individual-level output (option: empirical, strata: F x CH)
| Variable | Description |
|---|---|
EMPTH |
Empirically-determined threshold |
EMPF |
Relative frequency of above-thresholds points based on EMPTH |
MEAN_OVER_MEDIAN |
Ratio of mean to median, to index skewness of the wavelet coefficients |
Between-class variance over range of thresholds (option: empirical, strata: TH x F x CH)
| Variable | Description |
|---|---|
SIGMAB |
Between-class variance for given threshold |
Example
Here we use this approach on the three tutorial individuals: we run a
basic command to estimate spindles (for all NREM2 sleep, with no other
artifact detection in place), adding the empirical threshold:
luna s.lst sig=EEG -o out.db -s "MASK ifnot=NREM2 & RE & SPINDLES fc=11,15 empirical"
In lunaR, we use ldb() to load the resulting out.db file:
k <- ldb("out.db")
TH):
d <- lx( k , "SPINDLES" , "CH_F_TH" )
lid() and basic R functions to create three plots separately, of the between-class variance as a function of threshold, split by fast and slow spindles. First, we get a list of the individual IDs:
ids <- unique( d$ID )
par(mfcol=c(1,3))
for (id in ids) {
t <- lid( d , id )
plot( t$TH[ t$F==11 ] , t$SIGMAB[ t$F == 11 ] ,
type="l" , lwd=2 , col="orange" ,
main=id, xlab="Threshold" , ylab="Between-class variance" , ylim=c(0,1))
lines( t$TH[ t$F==15 ] , t$SIGMAB[ t$F == 15 ] , type="l" , lwd=2 , col="skyblue" )
legend( 5,0.2,c("11 Hz","15 Hz"),fill=c("orange","skyblue"))
}
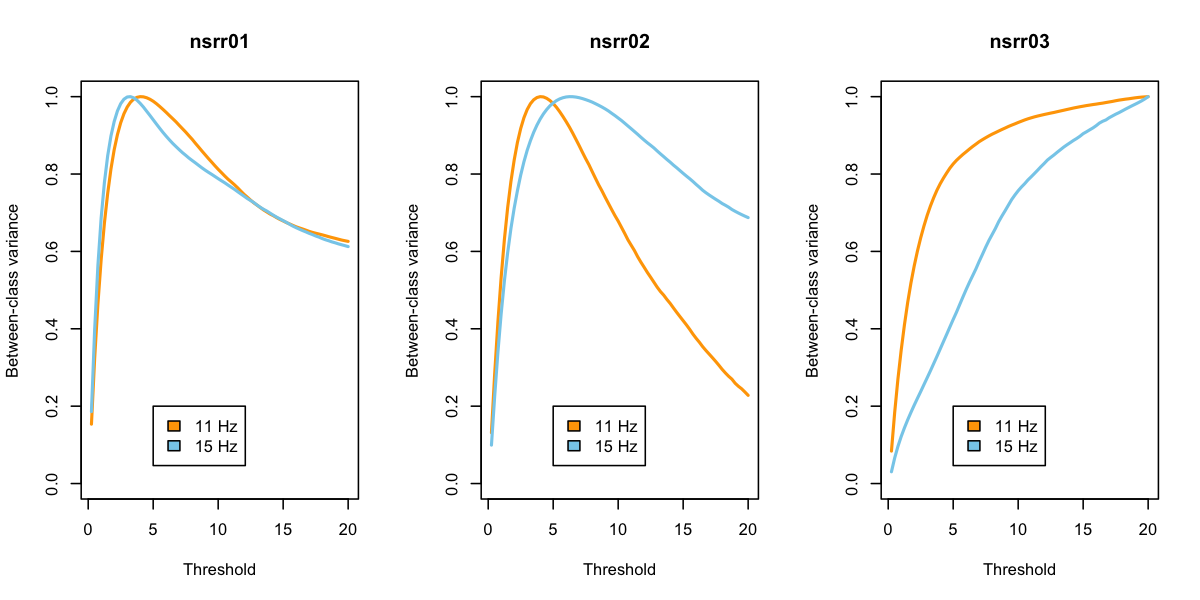
For the first two individuals, based on Otsu's criterion at least,
this suggests that thresholds around 4 or 5 are optimal, in the sense
of maximizing the between class variance (i.e. in wavelet coefficients
between "spindle" and "non-spindle" classes). For the third
individual, the results look strange, suggesting that extreme threshold
such as 20 times the mean is optimal. This is mirrored in the
individual-level output for EMPTH and EMPF: the optimal thresholds
and the corresponding area under the curve:
destrat out.db +SPINDLES -r F CH -v EMPTH EMPF -p 3
ID CH F EMPF EMPTH
nsrr01 EEG 11 0.034 4.000
nsrr01 EEG 15 0.067 3.250
nsrr02 EEG 11 0.041 4.000
nsrr02 EEG 15 0.024 6.250
nsrr03 EEG 11 0.001 20.000
nsrr03 EEG 15 0.002 20.000
This suggests there may be something off with the EEG channel for
nsrr03. As well as looking at the raw data, one approach may be to
impose some automated artifact detection prior to spindle detection
and threshold estimation (i.e. as we would typically recommend).
Re-running with aberrant epochs removed prior to spindle detection
(i.e. by including ARTIFACTS and
SIGSTATS commands:
luna s.lst sig=EEG -o out2.db -s "MASK ifnot=NREM2 & RE \
& ARTIFACTS mask & SIGSTATS mask th=3,3,3 & RE \
& SPINDLES fc=11,15 empirical"
This appears to normalize the "optimal" thresholds somewhat, in that the area under the curve (i.e. roughly corresponding to the portion of time in "spindle" versus "non-spindle" states) are ball-park what we may expect to observe in a typical individual who has around 2 or so spindles per minute detected at a central scalp EEG channel, with each spindle lasting approximately 1 second (i.e. 2/60 = 0.033):
destrat out2.db +SPINDLES -r F CH -v EMPTH EMPF -p 3
ID CH F EMPF EMPTH
nsrr01 EEG 11 0.046 3.500
nsrr01 EEG 15 0.092 2.750
nsrr02 EEG 11 0.041 4.000
nsrr02 EEG 15 0.029 5.500
nsrr03 EEG 11 0.033 4.000
nsrr03 EEG 15 0.066 3.000
Returning to the plots, things look a bit better:
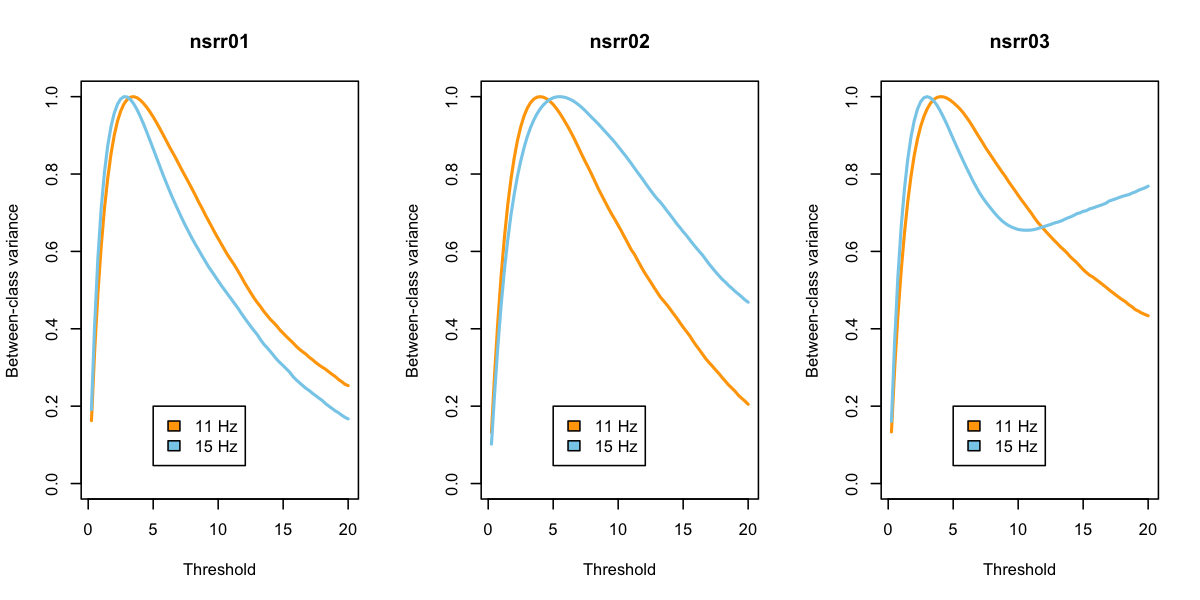
What to make of all this? We do not advise selecting different "optimal" thresholds for each individual based on these analyses. Rather, we suggest that looking at these (and other) metrics can be useful in basic sanity-checking for whether a given analysis for a given individual is likely to yield meaningful results.
Quality metrics
Luna calculates a simple QC metric for each spindle, by considering
the relative increase of non-spindle activity within the spindle
interval, relative to spindle-activity. We use five fixed bands,
which unlike the PSD command, are not altered by changing special
variables. The three
non-spindle bands are: delta (0.5-4 Hz), theta (4-8 Hz), and
beta (defined here as 20-30 Hz). The two spindle bands are slow
sigma (10-13.5 Hz) and fast sigma (13.5-16 Hz).
For each channel, Luna calculates the baseline power for each band, Pband. For the interval spanning the ith spindle, we calculate band power Piband. We then calculate relative enrichment for each band (on a log10 scale), as:
Eiband = log10(Piband) - log10(Pband).
The Q score for each spindle is based on the difference between the largest spindle-band relative enrichment versus the largest non-spindle band relative enrichment:
Qi = max( Eislow-sigma , Eifast-sigma ) - max( Eidelta , Eitheta , Eibeta )
That is, numbers less than zero indicate that a non-spindle band is showing greater relative enrichment (relative to the baseline log-scaled power for that band) than either spindle band. Such spindles are more likely to reflect artifact which had a non-specific effect on the power spectrum.
To illustrate this QC metric, we'll detect spindles for the second tutorial individual:
luna s.lst 2 sig=EEG -o out.db \
-s "MASK ifnot=NREM2 & RE & \
SPINDLES fc=11,15 enrich q=-9 annot=sp1 cycles=12"
Here, we detect both fast and slow spindles, targeting 11 and 15 Hz
respectively; we've set cycles to 12 instead of the default 7, in
order to increase frequency-specificity. We've added the enrich option to output
Eislow-sigma
and
Eifast-sigma
along with
Eidelta
Eitheta
and
Eibeta
for each spindle/channel/target-frequency/band (and so stratified by
CHxFxSPINDLExB). We also set q equal to -9, which effectively turns off any
QC filtering, so that we can see what some spindles that would not otherwise have been called
with the default threshold (q=0) look like. Finally, the ftr option writes out two files
that detail where spindles occur.
In lunaR:
library(luna)
lattach( lsl( "s.lst" ) , 2 )
ladd.annot.file( "id_nsrr02_feature_SPINDLES-spindles-EEG-wavelet-11-sp1.ftr" )
ladd.annot.file( "id_nsrr02_feature_SPINDLES-spindles-EEG-wavelet-15-sp1.ftr" )
out.db:
k <- ldb( "out.db" )
d <- lx( k , "SPINDLES" , "F" , "CH" , "SPINDLE" )
par(mfcol=c(1,2))
hist( d$Q[ d$F == 11 ] , breaks=30 , main="11 Hz spindles" , xlab="Q" , xlim=c(-2,2) )
abline(v=0,col="red",lwd=2)
hist( d$Q[ d$F == 15 ] , breaks=30 , main="15 Hz spindles" , xlab="Q" , xlim=c(-2,2) )
abline(v=0,col="red",lwd=2)
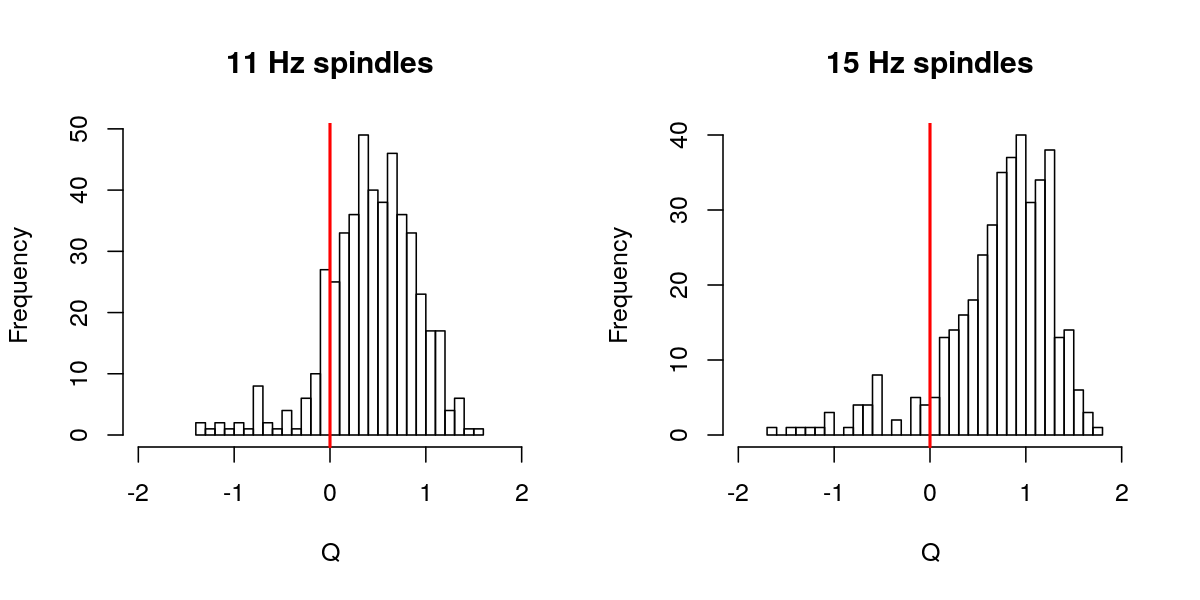
Of the full set (i.e. running without the default q=0 threshold in place), we see that approximately 10% of spindles have q less than 0:
table( d$Q > 0 , d$F )
11 15
FALSE 68 36
TRUE 405 370
Let's visualize some q<0 and q>0 spindles (we'll actually select
qc>1 spindles, to ensure that we don't randomly select spindles with
q very near 0). For simplicity, we'll do this just for fast spindles.
First, we'll save the labels of the two new annotation tracks we
appended (i.e. you can see the names from running lannots()):
a15 = "SPINDLES-spindles-EEG-wavelet-15-sp1"
s15 <- lannots( a15 )
length(s15) is indeed 406.
For visualization, here we'll just select five low Q and five high Q spindles:
lo15 <- sample(d$SPINDLE[ d$Q < 0 & d$F == 15 ])[1:5]
hi15 <- sample(d$SPINDLE[ d$Q > 1 & d$F == 15 ])[1:5]
Using lunaR functions, we can then write a function to plot the raw EEG signal for each spindle, with a 4-second flanking window on either side:
f1 <- function(i,a) {
x <- ldata.intervals( i , chs = "EEG" , annots = a , w = 4 )
y <- x$EEG
plot( x$SEC , y , type="l" , axes=F,xlab="",ylab="" , ylim=c(-125,125) )
t <- x$SEC[ x[,lsanitize(a)] == 1 ]
points( t , rep( -125,length(t)) , col=rgb(0,0,255,150,max=255) , pch="|" )
}
Finally, we'll generate a matrix of plots, with the five low Q spindles in the left column, and the five high Q spindles in the right column:
par(mfrow=c(5,2),mar=c(0,0,0,0) )
for (i in 1:5) {
f1( i = s15[ lo15[ i ] ] , a = a15 )
f1( i = s15[ hi15[ i ] ] , a = a15 )
}
As you can see, the low Q spindles (that would not have been
detected using Luna's default q=0 setting) appear to reflect some
sources of artifact, whereas, as a whole, the high Q spindles look
more like canonical spindles.
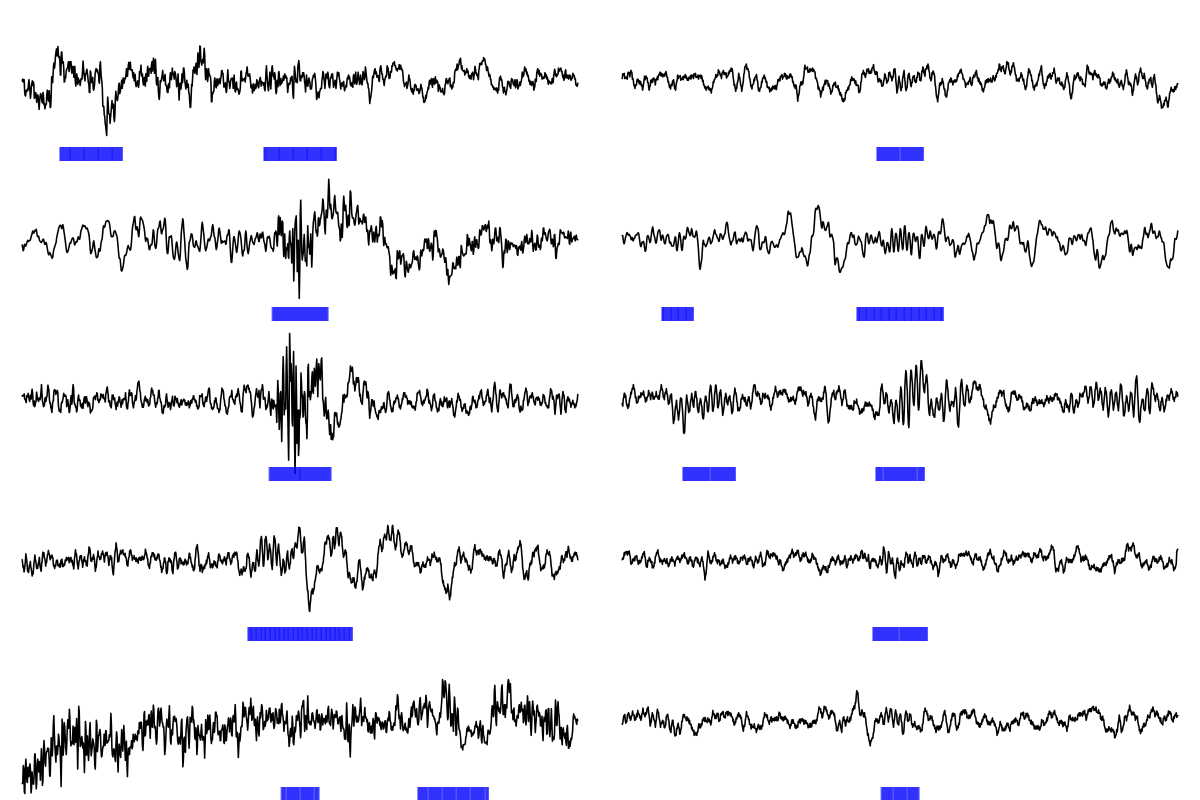
Be aware that setting q too high may unnecessarily miss many true
spindles though. A value of q=0.3 (i.e. log10(2)) requires that the
spindle-band enrichment is at least twice non-spindle band enrichment;
a value of q=1 requires a 10-fold enrichment. Considering the
distribution of mean Q over all individuals in the study may provide a good starting point,
if you want to select non-default values.
Warning
Naturally, neither this particular automated spindle detection algorithm, nor this particular quality metric, are guaranteed to always get it right... We fully expect that this algorithm will flag as a "spindle" events that most scorers would not, and vice versa. When first approaching this tool, we'd advise experimenting with the different thresholds and parameters, and spot-check studies in the way we have above. (The lunaR tutorial shows how to automatically iterate through all spindles detected.)
In general, if visual review seems to indicate likely artifact, the spindle is likely to be a false positive. In contrast, however, otherwise high Q spindles may appear relatively modest on visual review, and may not have been detected by some human scorers. Human scoring does not constitute a real gold-standard, however, and both human and automatic approaches impose inherently arbitrary thresholds on what is likely a continuously-varying and heterogeneous set of physiological processes. As such, we'd advise against automatically assuming the automatic detector is wrong, at least for this latter class of events ("low amplitude spindles").
Overall, the usual principles of a) trying to ensure that high-quality data go into the analysis, and b) working with large samples and appropriate statistical techniques downstream, are likely to be just as (if not more) important than spending too much tweaking the detection procedure, or agonizing over what is or is not "a true spindle".
Collating spindles
When detecting spindles on multiple channels or at multiple, nearby frequencies, what presumably is the same underlying physiological event may often be detected multiple times. These options assist in collating the results of spindle detection across channels and/or frequencies:
-
you can control whether two temporally close spindles detected on the same channel/target-frequency should be merged and considered as one event or not (
mergeparameter, the default is 0.5 seconds) -
second, within channel, you can collate temporally overlapping spindles detected at nearby frequencies (i.e. we would expect a great deal of overlap in the results of an analysis with FC=10 Hz and FC=11 Hz).
-
third, across channels, you can collate temporally overlapping spindles detected at nearby target frequencies; these may or may not represent physiologically distinct events depending on the montage and channels being collated.
These commands essentially take a list of spindles and generate another list of so-called m-spindles: groups of spindles that, on some definition, "overlap", i.e. represent the same underlying event.
The primary output from this analysis is an estimate of m-spindle
density (MSP_DENS), i.e. which is taken to reflect the total number
of unique spindles (i.e. allowing for potentially overlapping
definitions). For each m-spindle (set a spindles), Luna also
calculates an mean frequency (a weighted mean of the constituent
spindle frequencies, with weights given by the ISA of each spindle).
This is output for each spindle and also used to generate a
frequency-conditioned estimate of m-spindle density.
Parameters
| Parameter | Example | Description |
|---|---|---|
merge |
merge=0.2 |
Merge two putative spindles if within this interval (default 0.5 seconds) |
collate |
collate |
Within each channel, collate overlapping spindles with similar frequencies (to MSPINDLES) |
collate-channels |
collate-channels |
As above, except merge across channels also |
th-frq |
th-frq=1 |
Criterion for merging two spindles if difference in frequency is within this range (default 2 Hz) |
list-all-spindles |
list-all-spindles |
List all spindles that comprise each m-spindle |
Secondary parameters:
| Parameter | Example | Description |
|---|---|---|
th-interval |
th-interval=0.5 |
Merge two overlapping spindles if the ratio of intersection to union is at least this value (default 0, i.e. any overlap) |
th-interval-cross-channel |
||
th-interval-within-channel |
||
window |
window=0.5 |
Set window around each spindle when defining temporal overlap |
hms |
hms |
Show clock-time of each m-spindle |
Output
Individual-level summaries of m-spindles (option: collate, strata: none)
| Variable | Description |
|---|---|
MSP_DENS |
m-spindle density |
MSP_N |
m-spindle count |
MSP_MINS |
Denominator for density, i.e. minutes of signal analyzed |
m-spindle density stratified by m-spindle frequency (option: collate, strata: F )
| Variable | Description |
|---|---|
MSP_DENS |
m-spindle density conditional on m-spindle frequency |
Merged-spindle output (option: collate, strata: MSPINDLE)
or Merged-spindle output (option: collate-within-channel, strata: CH x MSPINDLE)
| Variable | Description |
|---|---|
MSP_DUR |
Duration of this m-spindle |
MSP_F |
Estimated frequency of this m-spindle |
MSP_FL |
Lower frequency of this m-spindle |
MSP_FU |
Upper frequency of this m-spindle |
MSP_SIZE |
Number of spindles in this m-spindle |
MSP_STAT |
Statistic for m-spindle |
MSP_START |
Start time (seconds elapsed from EDF start) of m-spindle |
MSP_STOP |
Stop time (seconds elapsed from EDF start) of m-spindle |
Spindle to m-spindle mappings (option: list-all-spindles, strata: SPINDLE x MSPINDLE)
| Variable | Description |
|---|---|
SCH |
Spindle label (channel:target frequency) |
FFT |
Spindle estimated frequency (via FFT) |
START |
Spindle start time (elapsed seconds from EDF start) |
STOP |
Spindle stop time (elapsed seconds from EDF start) |
Additional output (option: hms, strata: MSPINDLE or CH x MSPINDLE))
| Variable | Description |
|---|---|
MSP_START_HMS |
Merged spindle start clock-time |
MSP_STOP_HMS |
Merged spindle stop clock-time |
Example
luna s.lst 2 sig=EEG -o out.db -s "MASK ifnot=NREM2 & RE & \
SPINDLES fc-lower=10 fc-upper=16 fc-step=0.25 \
collate list-all-spindles"
The total m-spindle density is 4.89 spindles per second, i.e. this estimates the total unique number of spindles over this broad frequency range ( with target frequencies of 10-16 Hz and each wavelet spanning approximately plus/minus 2 Hz):
destrat out.db +SPINDLES
ID MSP_DENS MSP_MINS MSP_N
nsrr02 4.8972 199.5 977
We can also view the m-spindle density as a function of the
estimated m-spindle frequency: i.e. these numbers will sum to the
total MSP_DENS estimate (or close to it, as we only consider the
fixed range of 8 to 16 Hz in 0.5 Hz bins, by default, when outputing
this):
destrat out.db +SPINDLES -r F
Plotting the output of this command, we potentially see evidence of a bimodality in spindle frequencies for this individual:
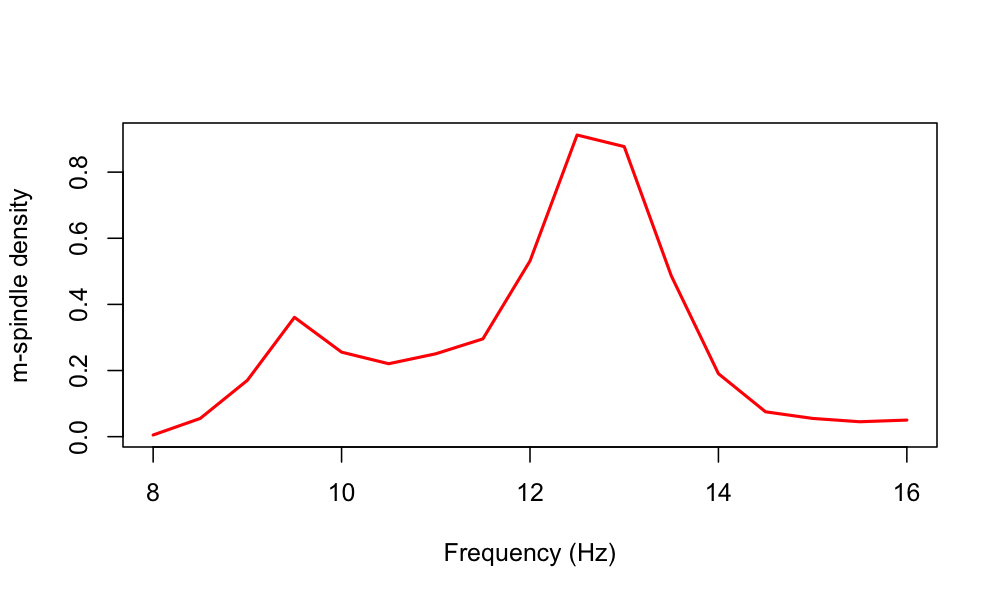
Relationship to individualized spindle detectors
Rather than using fixed target frequencies (e.g. 13.5 Hz, 11 Hz, or 15 Hz, etc) to define "spindles", some approaches to spindle detection seek to find, empirically, the "peak" or "peaks" (which may or may not be obviously present in the power spectra) for each particular individual, and then target spindles of those particular frequencies for that individual. In contrast, this approach is effectively agnostic to an individual's "true" characteristic spindle frequency (or frequencies), or to having to define spindle frequencies up front, by virtue of simply considering all possible frequencies in the broad sigma range, and then combining results. (Arguably this approach might be preferred a priori in any case, on the grounds that even within a single individual we can see marked shifts in spindle frequency over the course of the night.)
Other metrics
The following options report some additional spindle morphology
characteristics (these are unlikely to be desired as part of any
primary spindle analysis, and so can be ignored by most users). The
if option estimates spindle instantaneous frequency, averaged over
spindles, as a function of the relative location within the spindle.
The tlock option produces an averaged EEG signature of detected
spindles, synced to the spindle peak (point of max peak-to-peak
amplitude).
Parameters
| Parameter | Example | Description |
|---|---|---|
if |
if |
Estimate instantaneous frequency of spindles |
if-frq |
if-frq=1 |
Window around target frequency (default 2 hz) |
if-emp-frq |
if-emp-frq=1 |
Window around observed frequency (default 2 hz) |
tlock |
tlock |
Flag to request (verbose) average, peak-locked waveforms |
Instantaneous frequency is estimated via the filter-Hilbert method,
where the filter is FC +/- H Hz (where H is
set by if-frq, default 2). Using if-emp-frq over if-frq is typically
more robust and is preferred, however.
Output
Instantaneous frequency (IF) per spindle (option: if, strata: CH x F x SPINDLE)
| Variable | Description |
|---|---|
IF |
Mean frequency per spindle over duration |
Mean IF stratified by relative location in spindle (option: if, strata CH x F x RELLOC)
| Variable | Description |
|---|---|
IF |
Mean frequency of all spindles, per relative position within the spindle (five bins) |
Example
With the tutorial data:
luna s.lst 2 -o out.db -s "MASK ifnot=NREM2 & RE & \
SPINDLES sig=EEG fc=11,15 if tlock q=0.3 cycles=12"
FRQ and FFT,
as they are all estimating essentially the same thing):
destrat out.db +SPINDLES -r F CH SPINDLE -v FFT FRQ IF
To get the mean IF stratified by relative position with the spindle (i.e. dividing each spindle into five, equally-sized time bins):
destrat out.db +SPINDLES -r F CH RELLOC -p 3
ID CH F RELLOC IF
nsrr02 EEG 11 1 11.321
nsrr02 EEG 11 2 11.218
nsrr02 EEG 11 3 11.041
nsrr02 EEG 11 4 10.912
nsrr02 EEG 11 5 10.818
nsrr02 EEG 15 1 14.287
nsrr02 EEG 15 2 14.119
nsrr02 EEG 15 3 13.959
nsrr02 EEG 15 4 14.015
nsrr02 EEG 15 5 14.181
To plot the mean signal time-locked to each spindle peak (in lunaR):
k <- ldb("out.db")
d <- lx( k , "SPINDLES" , "CH" , "F" , "MSEC" )
par( mfcol=c(1,2) )
for (f in c(11,15)) {
plot( d$MSEC[ d$F==f ] , d$TLOCK[ d$F==f ] , ylim=c(-20,20) , type="l"
xlab="Time (sec)",ylab="Mean EEG",main=paste(f,"Hz spindles"), lwd=2)
abline(h=0,col="red",lty=2)
}
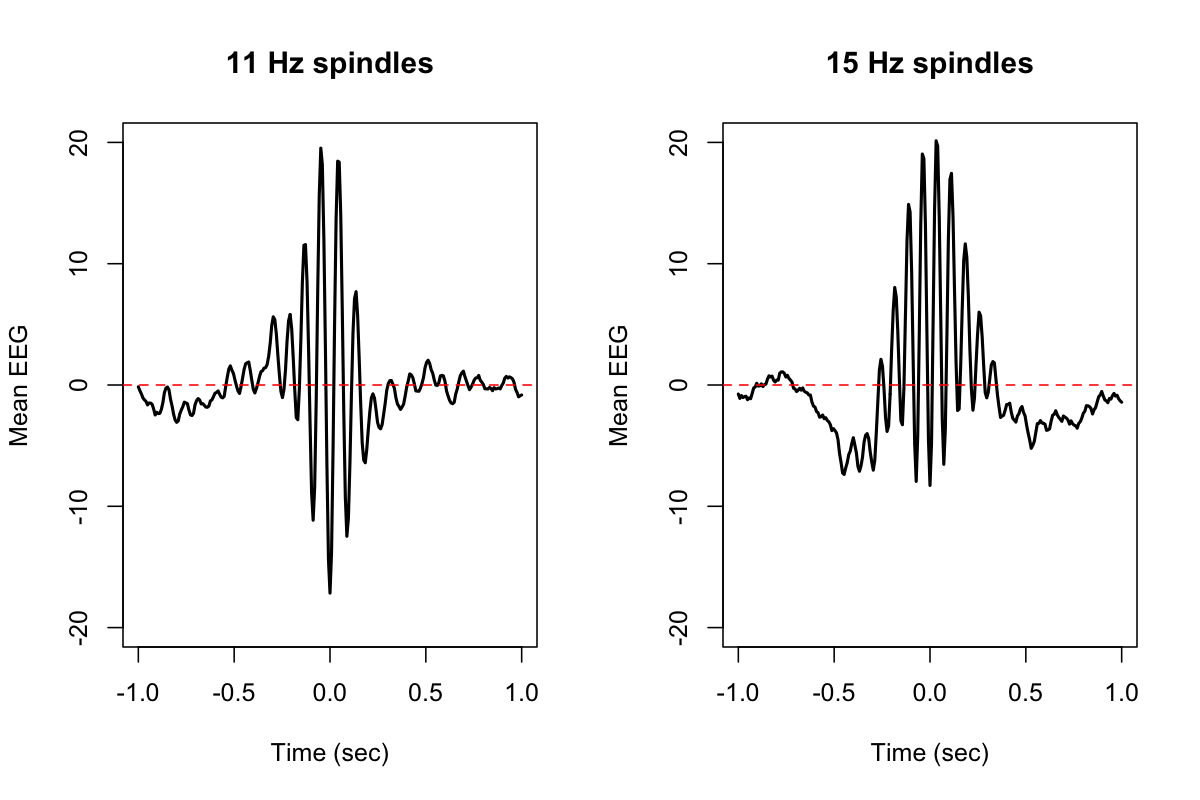
Annotations
Luna can generate annotation files representing the spindles detected in
a given run. These can be output (e.g. with WRITE-ANNOTS) and subsequently be attached to an EDF via lunaR, for example,
in order to visualize spindles, as in the examples above.
Parameters
| Parameter | Example | Description |
|---|---|---|
annot |
s1 |
Add an annotation to the in-memory dataset |
Output
The annot option generates annotations with the class ID as
specified by annot. The instance ID will be set to the target frequency; the
annotation channel ID will be set to the appropriate channel.
SPINDLES sig=C3,C4 fc=11,15 annot=sp1 & WRITE-ANNOTS file=sp1.annot
# sp1 | Spindle intervals
class instance channel start stop meta
sp1 15 C3 11861.797 11862.469 .
sp1 15 C4 11861.852 11862.531 .
sp1 15 C3 11866.898 11867.664 .
sp1 11 C3 11876.195 11876.984 .
sp1 11 C4 11876.195 11877.062 .
...
Spindle/SO coupling
Adding the so option for the SPINDLES command instructs Luna to
detect slow oscillations and report on their temporal coupling with
spindles. In short, Luna lines up the phase of slow oscillations with
the "peaks" of spindles (the point of maximal peak-to-peak amplitude,
typically near the center of the spindle), and then estimates the mean
SO angle across spindle peaks, and two metrics of that measure the
strength of non-random temporal coupling between SO and spindles.
Either all spindles all are included in this analysis (if the
all-spindles option is given), or only spindles that overlap a
detected SO (the default). In the former case, Luna will still use
the SO phase whether or not it would have explicitly reported a SO
spanning that spindle.
A magnitude metric (MAG) is calculated based on the intra-trial
phase clustering (ITPC) statistic for all spindles, or those
overlapping a detected SO. Second, an overlap metric (OVERLAP)
is calculated, reflecting the proportion of spindles that show any
overlap (temporally) with a SO (for that same channel). This second,
overlap metric is not reported if all-spindles is given.
If nreps is specified, Luna will perform that number of randomized
analyses, based on datasets in which spindle peaks are shuffled under
the null hypothesis of no specific coupling between spindles and SO.
The behavior of the permutation can be changed, such that instead of
spindles being shuffled only within the spanning epoch (the default),
they are shuffled across all epochs (perm-whole-trace). Here, the
entire set of spindle peaks is shifted by a constant (wrapping around
the end/start of the signal, as necessary). Second, if all-spindles
option is not specified, then when evaluating the magnitude of
coupling, spindles are shuffled (each independently) only within the
spanning SO (again, wrapping as necessary). That is, in this latter
case, each null replicate contains exactly the same number of spindles
and slow oscillations, the same number of which show overlap in each
replicate. The only thing that is randomized in this scenario is the
precise temporal association with spindle peak and SO phase within
detected SOs.
Parameters
For the primary parameters of the SO detection heuristic, see the section
on the SO command below. These include:
- frequency band (
f-lwr,f-upr, etc) - amplitude criteria (
mag,uV-neg,uV-p2p, etc) - duration criteria (
t-lwr,t-upr, etc)
In addition to the above, the so option of the SPINDLES command
has additional parameters for the analysis of spindle/SO coupling:
| Parameter | Example | Description |
|---|---|---|
nreps |
nreps=10000 |
Number of shuffled/surrogate time-series to generate |
all-spindles |
Consider all spindles, whether or not they overlap a SO | |
perm-whole-trace |
Do not use within-epoch shuffling | |
stratify-by-phase |
Additional overlap statistics per 20-degree SO phase bin |
Output
The so option of the SPINDLES command produces the same set of
outputs as the SO command (see below), describing the
number and properties of the detected SOs. In addition, the so
option of the SPINDLES command generates extra outputs describing
the temporal coupling between spindles and SOs, described here:
Primary individual-level spindle/SO coupling output (option: so, strata: CH x F)
| Variable | Description |
|---|---|
COUPL_MAG |
Magnitude of coupling (ITPC metric) |
COUPL_OVERLAP |
Number of spindles overlapping a detected SO |
COUPL_ANGLE |
Circular mean of SO phase at spindle peak (SO_SPHASE_PEAK below) |
COUPL_PV |
Asymptotic p-value for the ITPC statistic |
Permutation-based spindle/SO coupling output (options: so, nreps, strata: CH x F)
| Variable | Description |
|---|---|
COUPL_MAG_Z |
Z score for COUPL_MAG based on surrogate time series |
COUPL_MAG_NULL |
Mean coupling magnitude under the null |
COUPL_MAG_EMP |
Empirical p-value for COUPL_MAG based on surrogate time series |
COUPL_OVERLAP_NULL |
Mean overlap under the null |
COUPL_OVERLAP_Z |
Z-score for COUPL_OVERLAP |
COUPL_OVERLAP_EMP |
Empirical p-value for COUPL_OVERLAP |
COUPL_SIGPV_NULL |
Rate of null replicates with a p<0.05 significant asymptotic ITPC |
Spindle-level spindle/SO coupling output (option: so, strata: CH x F x SPINDLE)
| Variable | Description |
|---|---|
SO_SPHASE_PEAK |
SO phase at spindle peak |
NEAREST_SO |
Time to nearest SO |
NEAREST_SO_NUM |
Number of nearest SO |
SO morphology (option: so verbose-coupling, strata: CH x PHASE)
| Variable | Description |
|---|---|
SOPL_EEG |
Phase-locked mean EEG value (as a function of SO phase, in 20-deg bins) |
SO morphology (option: so verbose-coupling, strata: CH x SP)
| Variable | Description |
|---|---|
SOTL_EEG |
Time-locked mean EEG value (as a function of sample-points before/after SO negative peak) |
Spindle/SO phase coupling (option: so verbose-coupling, strata: CH x F x PHASE)
| Variable | Description |
|---|---|
SOPL_CWT |
SO-phased locked mean spindle wavelet coefficient (18 20-deg bins) |
Spindle/SO phase coupling (option: so verbose-coupling, strata: CH x F x SP)
| Variable | Description |
|---|---|
SOTL_CWT |
Time-locked mean spindle wavelet coefficient |
Example
Here we consider spindle/SO coupling for the second individual from
the tutorial data. We'll only consider NREM2 sleep, for fast (15 Hz
target frequency) and slow (11 Hz target frequency) at default
settings. We achieve this by adding so to the SPINDLES command,
along with the definition of the SO (see below). Here we'll
use a relative/adaptive threshold of 1.5 times the baseline mean,
along with othe default settings (e.g. for the duration and frequency
of the SO).
luna s.lst nsrr02 -o out.db -s 'MASK ifnot=NREM2 & RE & SPINDLES fc=11,15 sig=EEG so mag=1.5'
As seen from the log/console, Luna now also detected slow oscillations (as the SO command would do):
detecting slow waves: 0.2-4.5Hz
- relative threshold 1.5x median
- full waves, based on consecutive positive-to-negative zero-crossings
25380 zero crossings, of which 4977 met criteria (thresholds (<x, >p2p) -14.7917 30.0472)
running Hilbert transform
You can see information about the number and properties of detected SOs as follows (nb. not all output shown):
destrat out.db +SPINDLES -r CH | behead
ID nsrr02
CH EEG
SO 4977
SO_AMP -27.9675273648175
SO_DUR 0.686249547920436
SO_P2P 47.9202863568751
SO_RATE 24.9473684210526
That is, 4977 SO were detected, or 24.9 per minute (SO_RATE), with
an average duration of 0.69 seconds (SO_DUR) i.e. around 1.4 Hz.
Note these are based on NREM2 sleep only, and this definition of SO is
quite liberal, i.e. most of NREM2 is determined as having a "slow
oscillation" under this particular definition, and on average the SO
only has a negative deflection of -28uV. See below for
alternate definitions.
In terms of coupling between the SO and spindles, we can look at the
output table stratified by spindle target frequency (F) and channel
(CH):
destrat out.db +SPINDLES -r F CH | behead
Here are the relevant results for slow spindles:
COUPL_ANGLE 324.240526870805
COUPL_MAG 0.133073212508123
COUPL_PV 0.206789334851279
COUPL_OVERLAP 89
N 249
Here are the same metrics for fast spindles:
COUPL_ANGLE 267.757606950599
COUPL_MAG 0.456849609921408
COUPL_PV 2.04211688177307e-13
COUPL_OVERLAP 140
N 393
Based on the asymptotic p-values (COUPL_PV), we see highly
significant spindle/SO coupling for fast but not slow spindles in this
individual. 140 (of 393) fast spindles overlap a detected SO. For fast
spindles, the mean SO phase angle at spindle peaks is 267 degrees. SO
phase angle is defined starting from a positive-to-negative
zero-crossing, as follows:
| Location | Phase angle |
|---|---|
| Positive-to-negative zero-crossing | 0/360 degrees |
| Negative peak | 90 degrees |
| Negative-to-positive zero-crossing | 180 degrees |
| Positive peak | 270 degrees |
We can also see, for each spindle, whether or not the spindle overlapped a SO, and if so, the SO phase angle:
destrat out.db +SPINDLES -r F CH SPINDLE -v SO_PHASE_PEAK > o.txt
Looking at the results in R:
d <- read.table("o.txt",header=T)
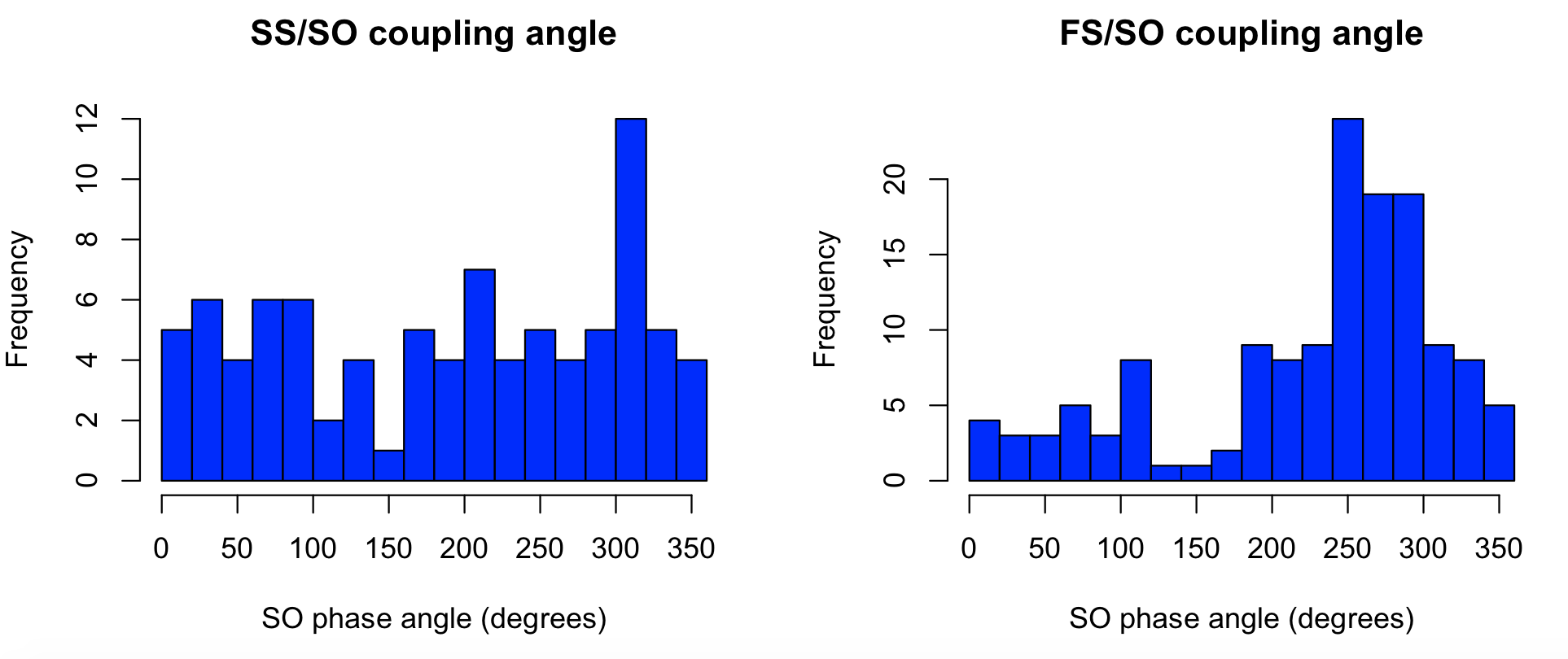
We see that fast spindles (FS) preferentially have their peak around the positive peak (270 degrees) of SO. In contrast, the pattern is less clear for slow spindles (SS) in this particular individual/analysis:
par(mfcol=c(1,2))
hist( d$SO_PHASE_PEAK[ d$F == 11 ] , breaks=15 , col = "blue" , xlab="SO phase angle (degrees)" , main="SS/SO coupling angle" )
hist( d$SO_PHASE_PEAK[ d$F == 15 ] , breaks=15 , col = "blue" , xlab="SO phase angle (degrees)" , main="FS/SO coupling angle" )
Next, we'll add random shuffling to the coupling analyses, to generate empirical, non-parametric estimates of significance, by adding the nreps command. Here we specify 100,000 random permutations per analysis (these are evaluated quite quickly), running now only for fast spindles:
luna s.lst nsrr02 -o out.db \
-s 'MASK ifnot=NREM2 & RE \
& SPINDLES fc=15 sig=EEG so mag=1.5 nreps=100000'
We now see some additional metrics in the output. For example,
whereas as observed 140 spindles overlapping a SO (COUPL_OVERLAP),
we see that on average, in each null replicate, only 104.2 spindles
overlapped a SO (COUPL_OVERLAP_NULL). Based on these statistics,
this gives an empirical p-value of 0.028 (COUPL_OVERLAP_EMP),
i.e. in this individual, we see significant (p<0.05) overlap between
spindles and SO at the gross level:
destrat out.db +SPINDLES -r F CH | behead
COUPL_ANGLE 267.757606950599
COUPL_MAG 0.456849609921408
COUPL_MAG_EMP 9.99990000099999e-06
COUPL_MAG_NULL 0.105944405707361
COUPL_MAG_Z 7.14893183381634
COUPL_OVERLAP 140
COUPL_OVERLAP_EMP 0.027649723502765
COUPL_OVERLAP_NULL 104.1836
COUPL_OVERLAP_Z 2.22339252291559
COUPL_PV 2.04211688177307e-13
COUPL_SIGPV_NULL 0.2081
The corresponding values for the ITPC metric are 0.46 observed
(COUPL_MAG) versus an average of 0.11 (COUPL_MAG_NULL) under the
null, giving an empirical p-value of 9.9e-6. Note: this is the
smallest possible empirical p-value if only 100,000 replicates are
specified, i.e. 1/(1+100000).
Note
As they are based on randomization, naturally, you may get numerically slightly different values for some of these metrics, if you repeat the above analysis on the tutorial data.)
Note that COUPL_SIGPV_NULL shows that over 20% of null-replicates
had an asymptotic ITPC p-value significant at the p<0.05 level.
That this is approximately four times greater than we'd expect by
chance suggests that the asymptotic p-values are, in this situation,
anti-conservative (i.e. we'd expect a 5% rate to be p<0.05 under the
null). The observed 2e-13 asymptotic p-value is therefore likely to
be inflated too. These considerations motivate the use of empirical
p-values in this context, given known biases with the ITPC (and
similar) metrics. That is, prefer to use COUPL_MAG_EMP or
COUPL_MAG_Z as measures of spindle/SO coupling strength, rather than
COUPL_MAG or COUPL_PV.
Next, we can run the same analysis, but adding the all-spindles
option to extend the coupling analysis to all spindles, i.e. whether
or not they overlap a detected SO:
luna s.lst nsrr02 -o out2.db -s 'MASK ifnot=NREM2 & RE & SPINDLES fc=15 sig=EEG so mag=1.5 nreps=100000 all-spindles'
Now the COUPL_OVERLAP metrics are not shown, i.e. as this analysis
ignores whether or not a particular SO was detected; rather the
estimated instantaneous phase from the Hilbert transform of the
slow-wave band-pass filtered signal is used irrespective:
COUPL_ANGLE 249.957585228371
COUPL_MAG 0.23828160431191
COUPL_MAG_EMP 0.010469895301047
COUPL_MAG_NULL 0.0555142478440371
COUPL_MAG_Z 4.60828305383158
COUPL_PV 2.03816238295918e-10
COUPL_SIGPV_NULL 0.13594
N 393
Here we see an attenuated but still significant spindle/SO coupling
(i.e. COUPL_MAG_EMP is 0.01). In this example, however, using the
larger number of spindles (i.e. all 393 here, rather than only the 140
SO-overlapping ones) does not appear to give stronger results
(although, not the asymptotic p-values are high in each case).
Verbose coupling output
The verbose-coupling option, along with so, produces some additional tables/output strata that can be used to generate plots describing the nature of spindle/SO coupling:
luna s.lst nsrr02 -o out.db -s 'MASK ifnot=NREM2 & RE & SPINDLES fc=15 sig=EEG so mag=1.5 nreps=100000 verbose-coupling'
Four additional strata are added to the output:
[SPINDLES] : CH PHASE : 18 level(s) : SOPL_EEG
: : :
[SPINDLES] : CH SP : 251 level(s) : SOTL_EEG
[SPINDLES] : F CH PHASE : 18 level(s) : SOPL_CWT
: : :
[SPINDLES] : F CH SP : 251 level(s) : SOTL_CWT
The first two contain the average EEG, either as a function of SO
phase, or time-locked to the negative peak of each SO. The second two
contain the average spindle wavelet power (and so are dependent on
F, the target frequency for the spindle analysis too). For example, we can extract and
use R to plot SOPL_EEG and SOPL_CWT, the SO phase-locked raw EEG and spindle CWT respectively:
par(mfcol=c(1,2))
plot( d$PHASE , d$SOPL_CWT , type="b" , xlab="SO phase angle (degrees)" , ylab="CWT coefficient" , pch=20 )
plot( d$PHASE , d$SOPL_EEG , type="b" , xlab="SO phase angle (degrees)" , ylab="Mean raw EEG (uV)" , pch=20 )
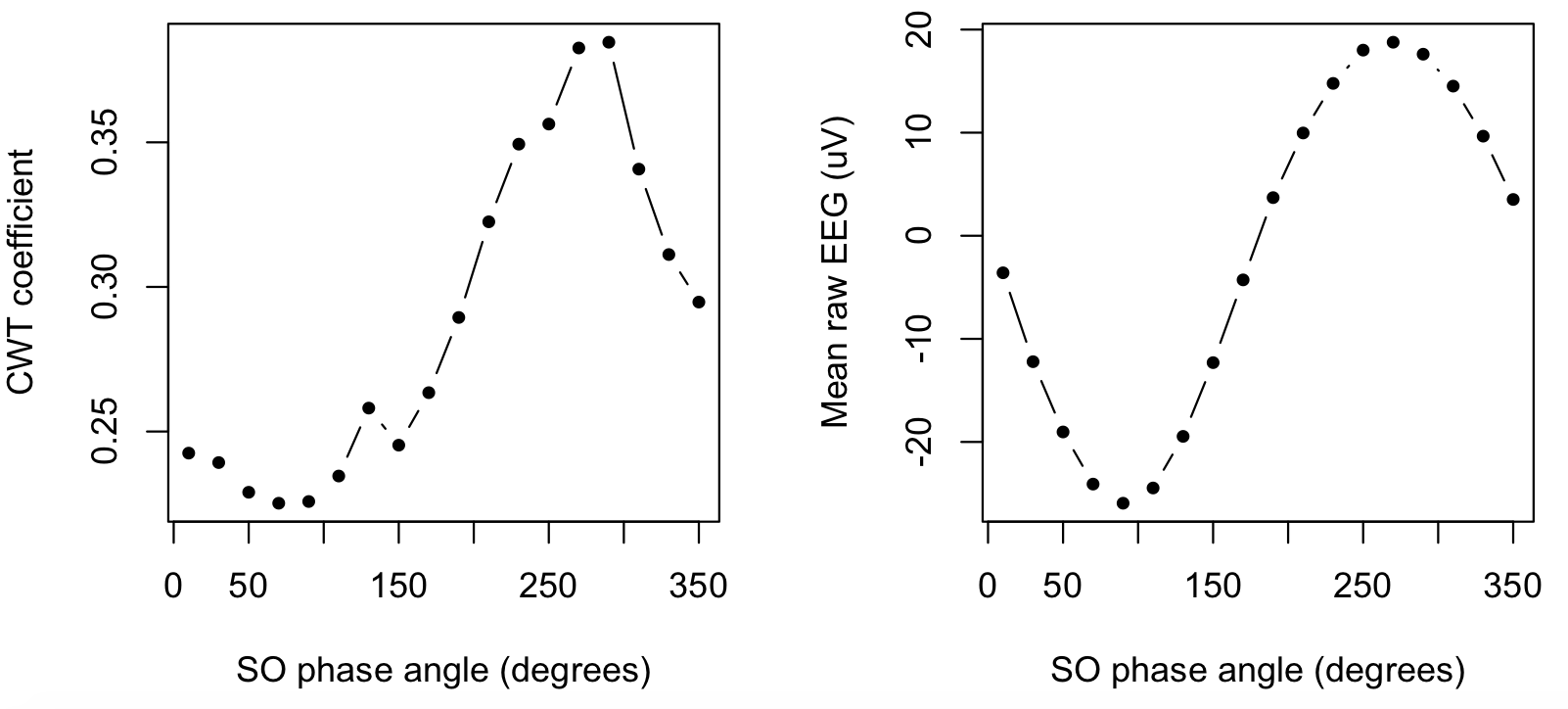
That is, the right panel confirms (as expected/defined) the typical SO, which has a positive peak at 270 degrees. The left panel shows how the spindle wavelet power (i.e. the underlying metric upon which spindles are explicitly detected) also has a peak that corresponds to the SO positive peak.
Second, we can request additional COUPL_OVERLAP output, where rather
than assessing any overlap between a spindle and a SO, here we
instead count overlap between a spindle and a particular point of a
SO, defined by SO phase (in 18 bins each of 20 degrees). That is, we
add the stratify-by-phase option:
luna s.lst nsrr02 -o out.db -s 'MASK ifnot=NREM2 & RE & SPINDLES fc=15 sig=EEG so mag=1.5 nreps=100000 stratify-by-phase'
This has the effect of generating an additional table/output strata, defined by F and PHASE for a given channel (CH):
[SPINDLES] : F CH PHASE : 18 level(s) : COUPL_OVERLAP COUPL_OVERLAP_EMP
: : : COUPL_OVERLAP_Z
The definitions of the COUPL_OVERLAP metrics are as above, except now they are conditional on SO-phase bin.
destrat out.db +SPINDLES -r F CH PHASE
ID CH F PHASE COUPL_OVERLAP COUPL_OVERLAP_EMP COUPL_OVERLAP_Z
nsrr02 EEG 15 10 4 0.934 -1.235
nsrr02 EEG 15 30 3 0.958 -1.378
nsrr02 EEG 15 50 3 0.958 -1.384
nsrr02 EEG 15 70 5 0.794 -0.637
nsrr02 EEG 15 90 3 0.981 -1.676
nsrr02 EEG 15 110 8 0.440 0.287
nsrr02 EEG 15 130 1 1.000 -2.450
nsrr02 EEG 15 150 1 1.000 -2.587
nsrr02 EEG 15 170 2 0.996 -2.047
nsrr02 EEG 15 190 9 0.447 0.261
nsrr02 EEG 15 210 8 0.642 -0.230
nsrr02 EEG 15 230 9 0.516 0.084
nsrr02 EEG 15 250 24 0.000 5.147
nsrr02 EEG 15 270 19 0.005 2.968
nsrr02 EEG 15 290 19 0.002 3.344
nsrr02 EEG 15 310 9 0.355 0.508
nsrr02 EEG 15 330 8 0.458 0.241
nsrr02 EEG 15 350 5 0.810 -0.685
That is, COUPL_OVERLAP still adds to 140, as in the above example,
but these spindles are now listed depending on exactly when in the
SO the spindle peak lies. Plotting these metrics in R:
par(mfrow=c(2,2))
plot( d$PHASE , d$COUPL_OVERLAP , pch=20 , type="b" , xlab="SO phase angle (degrees)" , ylab="N spindles overlapping SO" )
plot( d$PHASE , d$COUPL_OVERLAP_Z , pch=20 , type="b" , xlab="SO phase angle (degrees)" , ylab="Z statistic" )
plot( d$PHASE , d$COUPL_OVERLAP_EMP , pch=20 , type="b" , xlab="SO phase angle (degrees)" , ylab="Empirical p-value" )
plot( d$PHASE , -log10(d$COUPL_OVERLAP_EMP ), pch=20 , type="b" , xlab="SO phase angle (degrees)" , ylab="-log10(empirical p-value)" )
abline( h=-log10( 0.05) , lty=2)
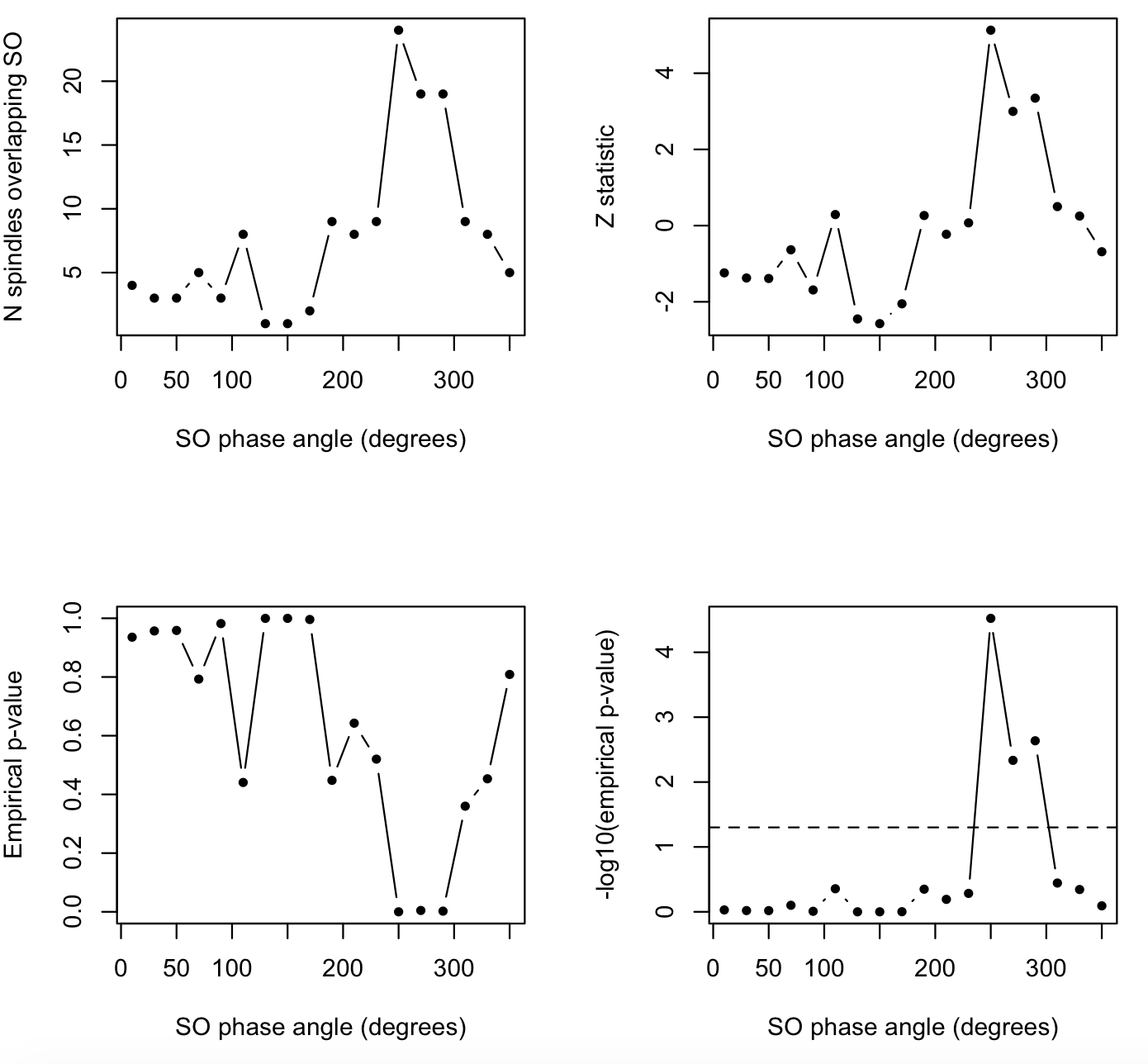
That is, we see the above-chance rate of spindle/SO overlap is not
uniformly distributed across the entire, but is concentrated around
250-290 degrees (consistent with the significant COUPL_MAG which is
another way to tell us the same thing).
See the SO section below for more details on how to define slow oscillations.
Misc
show-coef
The show-coef is primarily for development purposes and should not
be part of a standard spindle-analysis workflow. It produces a lot
of output: five values for every time-point for every channel for
every spindle frequency: this will be hard work for the output
database, which is not really designed for this type of data: you are
better off using this without the -o database option; rather,
extract the desired information directly from standard output:
luna s.lst 1 < cmd.txt \
| awk ' $2 == "SPINDLES" && $5 == "CWT" { print $3,$4,$6 } ' OFS="\t"
| Variable | Description |
|---|---|
RAWCWT |
Absolute, raw wavelet coefficient value |
CWT |
Normalized wavelet coefficient value |
CWT_TH |
Wavelet threshold for core spindle |
CWT_TH2 |
Wavelet threshold for flanking spindle |
CWT_THMAX |
Wavelet max threshold for spindle |
SO
Detects slow oscillations via a simple, heuristic approach
The SO command uses an heuristic approach to detect slow
oscillations (SO) in the sleep EEG, combined with filter-Hilbert estimation
of SO phase. This command can also be invoked through the so parameter of
SPINDLES command, in which case additional metrics
describing spindle/SO coupling are also reported.
The SO command works by first bandpass-filtering the EEG (given the frequency parameters) and then applies a series of time and amplitude criteria to identify individual SOs. Either absolute (fixed) or adaptive (relative) amplitude thresholds can be specified.
Parameters
The example section below illustrates the application of these parameters, which determine what constitutes a detected SO.
Bandpass filtering prior to SO detection:
| Parameter | Example | Description |
|---|---|---|
f-lwr |
f-lwr=0.2 |
Bandpass filter lower transition frequency |
f-upr |
f-upr=4.5 |
Bandpass filter upper transition frequency |
Time criteria for SO detection:
| Parameter | Example | Description |
|---|---|---|
t-lwr |
t-lwr= |
Minimum duration of a SO |
t-upr |
t-upr= |
Maximum duration of a SO |
t-neg-lwr |
t-neg-lwr=0.5 |
Lower threshold for negative peak (seconds) |
t-neg-upr |
t-neg-lwr=2 |
Upper threshold for negative peak (seconds) |
Amplitude criteria for SO detection:
| Parameter | Example | Description |
|---|---|---|
mag |
mag=1 |
Use a relative amplitude threshold instead (see below) |
uV-neg |
uV-neg=-75 |
Negative peak amplitude threshold ( default 0, i.e. no threshold ) |
uV-p2p |
uV-p2p=140 |
Peak-to-peak amplitude (default 0, i.e. no threshold) |
th-mean |
th-mean |
Use mean (instead of median) for thresholding |
stats-median |
stats-median |
Use median (instead of mean) for reporting SO stats (not for epoch-level reports) |
Additional options;
| Parameter | Example | Description |
|---|---|---|
tl |
tl=sig1,sig2 |
Other signals to be summarized w.r.t. time-locked SO |
onset |
onset |
Time-lock to SO onset instead of negative peak |
pos |
pos |
Time-lock to SO positive peak instead of negative peak |
window |
window=1 |
Window around SO peak (default +/- 3 seconds) |
Outputs
Primary SO information per channel (strata: CH)
| Variable | Description |
|---|---|
SO |
Number of SO detected |
SO_RATE |
SO per minute |
SO_AMP |
Median amplitude (of negative peak) |
SO_P2P |
Median peak-to-peak amplitude |
SO_DUR |
Median SO duration |
SO_SLOPE |
Median slope (by default, SO_SLOPE_NEG2 |
additional metrics w/ verbose: |
|
SO_SLOPE_NEG2 |
Median slope (negative peak to negative-to-positive zero-crossing) |
SO_NEG_DUR |
Median negative peak duration |
SO_POS_DUR |
Median positive peak duration |
SO_SLOPE_NEG1 |
Median slope (positive-to-negative zero-crossing to negative peak) |
SO_SLOPE_POS1 |
Median slope (negative-to-positive zero crossing to positive peak) |
SO_SLOPE_POS2 |
Median slope (positive peak to positive-to-negative zero-crossing) |
SO_TH_NEG |
Threshold used for negative peak |
SO_TH_P2P |
Threshold used for peak-to-peak |
Per SO output (strata: CH x N)
| Variable | Description |
|---|---|
START |
Start of SO (in seconds elapsed from start of EDF) |
STOP |
Stop of SO (in seconds elapsed from start of EDF) |
DUR |
SO duration |
UP_AMP |
SO positive peak amplitude |
DOWN_AMP |
SO negative peak amplitude |
P2P_AMP |
SO peak-to-peak amplitude |
SLOPE_NEG2 |
Slope (upward slope of negative peak) |
| additional metrics: | |
START_IDX |
Sample index of SO start |
STOP_IDX |
Sample index of SO stop |
UP_IDX |
Sample index of positive peak |
DOWN_IDX |
Sample index of negative peak |
SLOPE_POS1 |
Slope (alternate metric) |
SLOPE_POS2 |
Slope (alternate metric) |
SLOPE_NEG1 |
Slope (alternate metric) |
Epoch-level counts of SO (strata: CH x E)
| Variable | Description |
|---|---|
N |
Number of SOs detected in that epoch |
Time-locked signal averaging (option: tl, strata: CH x CH2 x SP)
| Variable | Description |
|---|---|
SOTL |
Average signal for CH2, time-locked by SO detected in CH |
Example
The SO detection routine operates as follows:
-
Band-pass filter the signal with transition frequencies
f-lwrandf-upr(defaulting to 0.5 and 4 Hz) -
Find all positive-to-negative zero-crossings in the filtered signal
-
Retain intervals between consecutive zero-crossings that are between
t-lwrandt-uprin duration (defaulting to 0.8 and 2.0 seconds) -
Either apply absolute amplitude filtering, designating SO as intervals with a negative peak below
uV-neg(e.g. ifuV-neg=-75this means more negative, i.e. -80 uV ) and a peak-to-peak amplitude greater thanuV-p2p -
Or apply adaptive amplitude filtering, designing SO as intervals with negative peak of
magtimes the median (or mean, ifth-mean) negative peak and a peak-to-peak amplitudemagtimes the median (or mean, ifth-mean) peak-to-peak amplitude; the thresholds are written to the output (SO_TH_NEGandSO_TH_P2P)
As an example, looking at N3 sleep in the second individual from the
tutorial dataset, for the first EEG channel (EEG),
and using an adaptive threshold with a factor of 2 times the median:
luna s.lst nsrr02 -o out.db -s 'MASK ifnot=NREM3 & RE & SO sig=EEG mag=2'
detecting slow waves: 0.2-4.5Hz
- relative threshold 2x median
- full waves, based on consecutive positive-to-negative zero-crossings
9418 zero crossings, of which 824 met criteria (thresholds (<x, >p2p) -34.1888 69.5302)
running Hilbert transform
Looking at the primary output variables:
destrat out.db +SO -r CH | behead
SO 824
SO_AMP -51.3357345906725
SO_DUR 1.0331359223301
SO_P2P 89.7069054223203
SO_RATE 8.90810810810811
We see there were 824 SOs detected, or a rate of 8.9 per minute. On average, each SO was 1.03 seconds long, had a negative peak amplitude of -51.3uV, and a peak-to-peak amplitude of 89.7uV.
In the literature, different investigators have used different
definitions for SOs in different contexts. Instead of using a relative
amplitude threshold (mag), you could set an absolute threshold. For
example, here we require a negative peak of at least -40 uV, and at
least 75 uV peak-to-peak:
luna s.lst nsrr02 -o out.db
-s 'MASK ifnot=NREM3 & RE & SO sig=EEG uV-neg=-40 uV-p2p=75'
SO 555
SO_AMP -56.1938081995994
SO_DUR 1.04608288288288
SO_P2P 96.1411853282356
SO_RATE 6
Alternatively, here we require even larger waveforms:
luna s.lst nsrr02 -o out.db
-s 'MASK ifnot=NREM3 & RE & SO sig=EEG uV-neg=-75 uV-p2p=140'
SO 13
SO_AMP -94.4252281038546
SO_DUR 1.08553846153846
SO_P2P 168.084348953554
SO_RATE 0.140540540540541
Naturally, the amplitude is larger here, but there are far fewer SOs (here fewer than one per minute).
Hint
It is also possible to specify time limits for the positive and negative components separately:
t-neg-lwr=0.3 t-neg-upr=1.5 t-pos-lwr=0.0 t-pos-upr=1.0
Here we combine all definitions in a single command, this time for N2
and N3 sleep combined, using TAG to organize the
results (by adding a tag R). We'll also generate data to visualize
the average waveforms, using tl:
luna s.lst nsrr02 -o out.db
-s 'MASK all & MASK unmask-if=NREM2,NREM3 & RE &
TAG R/1 & SO sig=EEG mag=2 tl=EEG window=2 &
TAG R/2 & SO sig=EEG uV-neg=-40 uV-p2p=75 tl=EEG window=2 &
TAG R/3 & SO sig=EEG uV-neg=-75 uV-p2p=140 tl=EEG window=2 '
Looking at the SO rate for each threshold definition, we recover the above values:
destrat out.db +SO -r R CH -v SO_RATE SO
ID CH R SO SO_RATE
nsrr02 EEG 1 4304 14.74
nsrr02 EEG 2 896 3.07
nsrr02 EEG 3 25 0.086
Now we see a larger difference in the estimated SO rate between the
first two methods (R of 1 and 2), reflecting that fact that adding
N2 sleep effectively lowers the threshold for detecting SOs, leading
to an overall higher rate, which may at first be counter-intuitive.
In contrast, for the two absolute definitions, the average rate lowers
when considering N2 and N3 combined versus just N3 sleep.
Extracting information from the tl option, this gives the averaged EEG signal time-locked to the negative SO peak:
destrat out.db +SO -r R CH CH2 SP > d.txt
d <- read.table("d.txt",header=T)
plot( d$SP[d$R==1], d$SOTL[d$R==1] , type="l" , col="blue" , lwd = 2 ,
ylim = c(-100 , 50 ) ,
xlab="Time from negative SO peak (sample-points)" , ylab="Signal" )
lines( d$SP[d$R==2], d$SOTL[d$R==2] , col="red" , lwd = 2 )
lines( d$SP[d$R==3], d$SOTL[d$R==3] , col="green" , lwd = 2 )
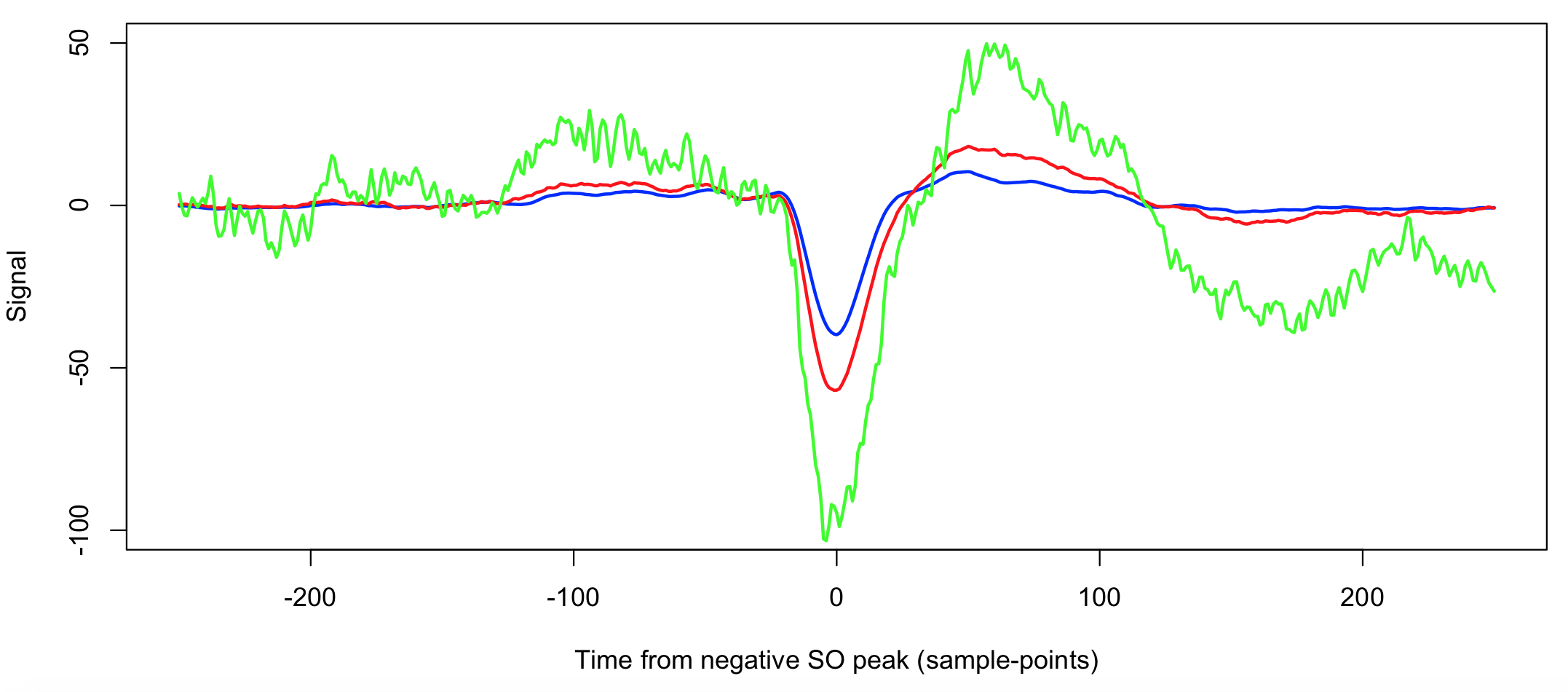
The three methods 1, 2 and 3 are blue, red and green respectively. The green line (R3) is more variable as a smaller number of SO were averaged over. Otherwise, we see that the second method (red line) yields slightly longer SOs of slightly greater amplitude.
Depending on the study population (e.g. children versus elderly, or mice versus human), and depending on the purpose of the analysis, different approaches may give better results. For example, estimating spindle/SO coupling will not be possible if there are not a sufficient absolute number of SOs (and/or spindles) in the recording.
luna s.lst nsrr02 -o out.db -s 'MASK all & MASK unmask-if=NREM2,NREM3 & RE &
TAG R/1 & SPINDLES so fc=15 sig=EEG mag=2 nreps=1000000 verbose-coupling &
TAG R/2 & SPINDLES so fc=15 sig=EEG uV-neg=-40 uV-p2p=75 nreps=1000000 verbose-coupling &
TAG R/3 & SPINDLES so fc=15 sig=EEG uV-neg=-75 uV-p2p=140 nreps=1000000 verbose-coupling '
destrat out.db +SPINDLES -r R CH F -v COUPL_OVERLAP COUPL_MAG_Z COUPL_OVERLAP_Z | behead
ID nsrr02
CH EEG
F 15
R 1
COUPL_MAG_Z 9.2884209252313
COUPL_OVERLAP 115
COUPL_OVERLAP_Z 2.99092347889109
ID nsrr02
CH EEG
F 15
R 2
COUPL_MAG_Z 6.49847282659681
COUPL_OVERLAP 54
COUPL_OVERLAP_Z 4.7739523530548
ID nsrr02
CH EEG
F 15
R 3
COUPL_MAG_Z 0
COUPL_OVERLAP 1
COUPL_OVERLAP_Z 0.770177278950557
Here we see greater evidence for non-random the magnitude of
spindle/SO coupling (based on the empirical distribution of the ITPC
statistic) under the first method (Z=9.2 for R1 versus Z=6.5 for R2),
likely due to the greater number of observations. The third method
(R3) only detects a couple of SOs in the dataset, with only a single
instance of spindle/SO overlap (COUPL_OVERLAP is 1). In terms of
gross overlap, R2 is slightly more power than R1 in this instance
(Z=4.8 for R2 versus Z=3.0 for R1).
destrat out.db +SPINDLES -r R F CH PHASE > d.txt
d <- read.table("d.txt",header=T)
plot( d$PHASE[d$R==1], d$SOPL_CWT[d$R==1],
type="l" , lwd=2, col="blue" , ylim=c(0,0.6) ,
xlab="SO phase (0/360 is positive-to-negative zero-crossing)" ,
ylab="Spindle wavelet power")
lines( d$PHASE[d$R==2], d$SOPL_CWT[d$R==2] , lwd=2, col="red" )
abline(v=270,lty=2)
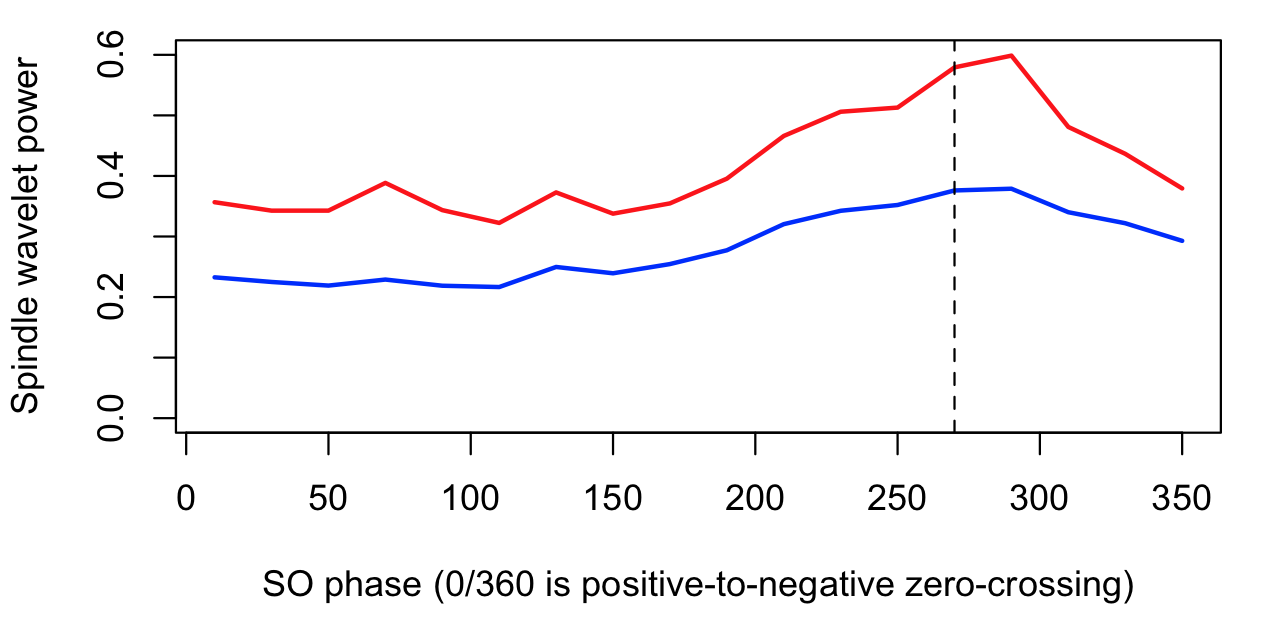
Here we see, for both R1 and R2 definitions, an increased fast spindle wavelet power around the negative peak of the SO (270 degrees) for this individual.