Cross-signal analyses
| Command | Description |
|---|---|
COH |
Pairwise channel coherence |
CORREL |
Pairwise channel correlation |
CC |
Coupling (dPAC) and connectivity (wPLI) |
PSI |
Phase slope index (PSI) connectivity metric |
XCORR |
Cross-correlation |
MI |
Mutual information |
TSYNC |
Cross-correlation and phase delay (alternate implementation) |
GP |
Granger prediction |
COH
Calculates pairwise spectral coherence across channels
The COH function calculates the magnitude-squared
coherence
for pairs of signals with similar sampling rates.
Parameters
| Parameter | Example | Description |
|---|---|---|
sig |
sig=C3,C4 |
An optional parameter, to specify which channels to calculate pairwise coherence between |
sig1 |
sig1=C3,C3 |
In stead of sig, specify all sig1 by sig2 channel pairs |
sig2 |
sig2=F3,F4 |
As above |
sr |
sr=125 |
Optionally, set sample rates to sr if different for a particular channel (i.e. to ensure that all signals have similar sampling rates |
spectrum |
spectrum |
Output full cross-spectra coherence rather than just in frequency bands (delta, theta, etc) |
epoch |
epoch |
A flag to output coherence statistics for per epoch as well as the whole signal |
epoch-spectrum |
epoch-spectrum |
A flag to output full cross-spectra coherence statistics for each epoch/channel pair |
Warning
If requesting the full spectrum and epoch-level analyses for a large number of channels, the output database may be very large... In this case, you should use
text-table mode output (with -t) for better performance when working with the output (as destrat can be slow othewise, for datasets with
very large numbers of strata).
Using sig1 and sig2 instead of sig can be more efficient, if you
are only interested in particular combinations of pairs. For example,
to see the pairwsie coherences between one channel and four others:
sig1=C3 sig2=F3,F4,O1,O2
sig=C3,F3,F4,O1,O2 would
evaluate 10 pairs, even if some pairwise comparisons are not of
interest).
Output
Coherence for power bands (B x CH1 x CH2)
| Variable | Description |
|---|---|
COH |
Magnitude-squared coherence |
ICOH |
Imaginary coherence |
LCOH |
Lagged coherence |
Full cross-spectra coherence (option: spectrum, strata: F x CH1 x CH2)
| Variable | Description |
|---|---|
COH |
Magnitude-squared coherence |
ICOH |
Imaginary coherence |
LCOH |
Lagged coherence |
Coherence for power bands, per epoch (option: epoch, strata: E x B x CH1 x CH2)
| Variable | Description |
|---|---|
COH |
Magnitude-squared coherence |
ICOH |
Imaginary coherence |
LCOH |
Lagged coherence |
Full cross-spectra, per epoch (option: epoch and spectrum, strata: E x F x CH1 x CH2)
| Variable | Description |
|---|---|
COH |
Magnitude-squared coherence |
ICOH |
Imaginary coherence |
LCOH |
Lagged coherence |
Example
To estimate the cross-spectra (up to 20 Hz) between the two EEG channels during NREM2
sleep for the tutorial individual nsrr02:
luna s.lst nsrr02 -o out.db -s "MASK ifnot=NREM2 & RE \
& COH spectrum max=20 sig=EEG,EEG(sec)"
Loading this into lunaR,
k <- ldb( "out.db" )
d <- lx(k,"COH","CH1","CH2","F")
plot( d$F, d$COH , type="l" ,
xlab="Frequency (Hz)" , ylab="Coherence" ,
ylim=c(0,1) , lwd=2, col="red")
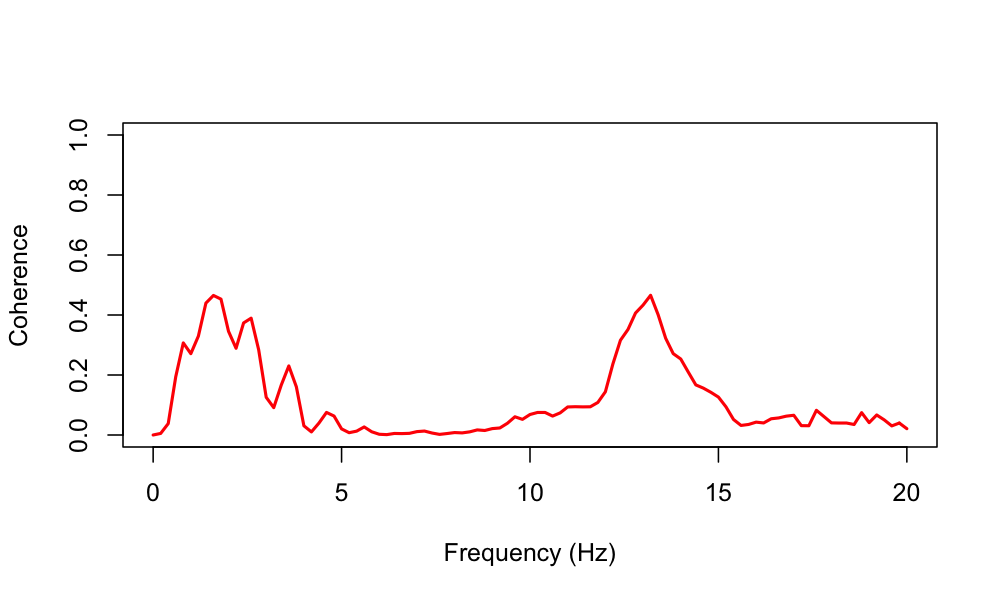
Following other reports (for example), we see peaks in EEG coherence especially for the low delta and sigma frequencies during NREM sleep. Naturally, the precise interpretation of coherence analysis (like any sleep EEG analysis) depends on the exact choice of montages, referencing and other technical and subject-level details, not to mention the potential for artifact (e.g. as illustrated here).
CORREL
Calculates pairwise Pearson correlation between signals
CORREL estimates pairwise correlation coefficients between signals,
either for the whole signal, or epoch-by-epoch. When epoch-level
statistics are requested, Luna also reports the mean and median of all
per-epoch statistics for a given channel pair.
As of v0.27, Luna also provides functions to incorporate spatial channel distance (i.e. for the EEG/MEG context) of channels, and to find disjoint sets of highly correlated channels.
Parameters
| Parameter | Example | Description |
|---|---|---|
sig |
sig=C3,C4,F3,F4 |
Optionally specify which signals to correlate (otherwise, do all) |
sig1 |
sig=C3,C4 |
Alternatively, only evaluate all sig1 by sig2 pairs |
sig2 |
sig=F3,F4 |
As above |
sr |
sr=128 |
Resample channels to this sample rate, if they are not already at that rate |
epoch |
epoch |
Display per-epoch correlations, and estimate mean and median correlation across epochs |
Epoch-level correlations
By default, the correlations from CORREL are based on the entire signal.
To instead estimate of channel-pair correlations based on
aggregating epoch-level correlations, use the ch-epoch option. This is often likely
more robust to artifacts. In particular, if you also use ch-median
then the final correlation is the median of epoch-level correlations
(otherwise, default = mean). In general, when using the CORREL command, it is probably
advisable to always add ch-epoch ch-median.
| Parameter | Example | Description |
|---|---|---|
ch-epoch |
Base overall correlations on aggregate summaries of epoch-level correlations | |
ch-median |
Use the median (instead of the mean) when applying ch-epoch |
Including EEG channel topographies
For high-density EEG/MEG studies, CORREL can produce some simple
metrics that flag pairs of channels that are more correlated by might
be expected given their topographical similarity. This may be indicative
of artifact or bridging, etc.
To include spatial distances in correlations, it is first necessary to
have previously attached a set of channel locations via the
CLOCS command) prior to running CORREL. An example map (for a
64-channel EEG) can be found
here: e.g.
CLOCS clocs=clocs64
CORREL sig=${eeg}
S variables from the CORREL command:
destrat out.db +CORREL -r CH1 CH2
S will obviously be the same of all individuals.)
ch-spatial-threshold ch-spatial-weight to CORREL,
Disjoint sets of highly correlated channels
CORREL now prints disjoint sets of highly-correlated channels (as
defined by the ch-high option) channels, with CHS stratifier, if
the ch-high option is given.
Output
Whole-signal correlations for pairs of channels (strata: CH1 x CH2)
| Variable | Description |
|---|---|
R |
Pearson product moment correlation |
R_MEAN |
(If epoch is specified) the mean of epoch-level correlations |
R_MEDIAN |
(If epoch is specified) the median of epoch-level correlations |
S |
If channel locations attached for this pair, the cosine similarity (based only on map) |
Whole-signal correlations for pairs of channels (option: epoch, strata: E x CH1 x CH2)
| Variable | Description |
|---|---|
R |
Per-epoch Pearson product moment correlation |
Channel-level summaries (strata: CH)
| Variable | Description |
|---|---|
SUMM_MEAN |
Mean correlation with all other channels |
SUMM_N |
Number of other channels this is correlated with |
SUMM_MIN |
Minimim correlation for this channel |
SUMM_MAX |
Maximum correlation for this channel |
SUMM_HIGH |
Number of correlations (i.e. channels) above ch-high |
SUMM_LOW |
Number of correlations (i.e. channels) below ch-low |
Disjoint sets of highly correlated channels (potion: ch-high, strata: none )
| Variable | Description |
|---|---|
SUMM_HIGH_N |
Total number of disjoint sets in the data |
SUMM_HIGH_CHS |
Comma-delimited list of channnels in this highly-correlated set |
Disjoint sets of highly correlated channels (option: ch-high, strata: CHS)
| Variable | Description |
|---|---|
N |
Number of channels in this disjoint set |
SET |
Channels in this disjoint set |
Example
Here we consider the epoch-by-epoch correlation between the two EOG
channels for tutorial subject nsrr02. Using
lunaC to estimate and report correlations at
the epoch level:
luna s.lst nsrr02 -o out.db -s "MASK if=wake & RE \
& STAGE \
& CORREL epoch verbose sig=EOG(L),EOG(R)"
STAGE
command, which we can use later when plotting results:
Using lunaR to examine the output (in R):
library(luna)
We'll attach the output database just created:
k <- ldb( "out.db" )
Examining the contents with lx():
lx(k)
CORREL : CH1_CH2 CH1_CH2_E
MASK : EPOCH_MASK
RE : BL
STAGE : E
First, we'll extract a simple vector of sleep stage (the STAGE variable from the STAGE command):
ss <- lx( k , "STAGE" , "E" )$STAGE
Second, we'll extract the epoch-by-epoch correlations:
d <- lx( k , "CORREL" , "CH1" , "CH2" , "E" )
Viewing data-frame d, note that the epochs (E) do not start at 1,
because we previously restricted this analysis to sleep epochs:
head(d)
ID CH1 CH2 E R
1 nsrr02 EOG(L) EOG(R) 92 0.5052592
2 nsrr02 EOG(L) EOG(R) 93 0.7998685
3 nsrr02 EOG(L) EOG(R) 98 0.4698097
4 nsrr02 EOG(L) EOG(R) 99 0.3147986
5 nsrr02 EOG(L) EOG(R) 100 0.3153311
6 nsrr02 EOG(L) EOG(R) 101 0.6581321
Finally, we can plot these epochs:
plot( d$E , d$R , col = lstgcols( ss ) , pch=20 , ylab="L-R EOG correlation", xlab="Epoch", ylim=c(-1,1) )
abline( h=0 , lty=2 )
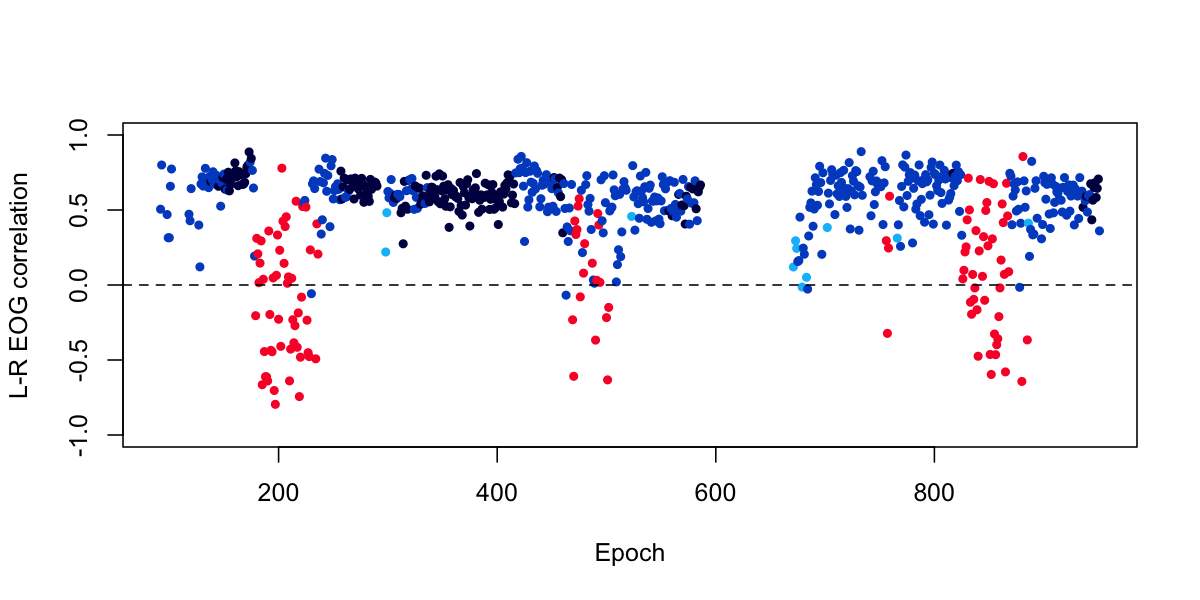
For this particular individual, there seems to be a clear and interesting pattern, in which we see that REM epochs (in red) are more likely to show negative correlations between the left and right EOG, reflecting the deviations due to eye-movements during REM.
Disjoint sets of highly correlated channels
CORREL now prints disjoint sets of highly-correlated channels (as defined by the ch-high option):
channels, with CHS stratifier, e.g.
destrat out.db +CORREL -r CHS
ID CHS N SET
ID01 1 2 Fp2,AF4
ID01 2 2 F3,F1
ID01 3 3 F2,FC2,FZ
ID01 4 6 C2,CP2,P2,P4,CPZ,PZ
ID01 5 2 CP4,CP6
ID01 6 2 P3,P1
ID01 7 2 P6,P8
ID01 8 4 PO4,O2,POz,OZ
i.e. here we have 8 groups (CHS), where N is the number of channels in
that group.
The baseline CORREL output will list the total # of channels with a highly-correlated partners:
destrat out.db +CORREL
ID SUMM_HIGH_CHS SUMM_HIGH_N
ID01 CPZ,P4 2
ID02 NA 0
ID03 NA 0
ID04 CP2,CP6 2
ID05 NA 0
ID06 NA 0
ID07 P3,TP7 2
ID08 P1,P2,P7,P8 4
ID09 CP2,O2,OZ,P4,POz 5
ID10 NA 0
ID11 AF4,C4,CP2,CP6,F2,F4,F6,FC2,FC4 9
ID11 P1,P7 2
ID12 AF4,FC4,P4,TP8 4
... etc ...
Spatial thresholding
Note that these statistics/sets are optionally influenced by
ch-spatial-threshold, which only includes channel pairs that are
below a certain similarity (i.e. not neighbouring). You can look at
the distribution of S from the output of +COREEL -r CH1 CH2 (see
above). Based on the above example map with 57 channels present in
the EDF, something like 0.8 excludes ~222 out of the ~1600 pairs,
i.e. on average, this will exclude the closest ~4 channels for each
channel (4 * 57 = 228). Valid arguments are between -1 and +1.
With spatial thresholding, channel pairs with a spatial similarity above threshold
are excluded from all correlational analyses. The column SUMM_N gives the number of
included channels after these step.
CORREL sig=${eeg} ch-epoch ch-spatial-threshold=0.8 ch-high=0.98 '
i.e.
luna s.lst -o out.db \
-s 'CLOCS file=clocs64
REFERENCE sig=${eeg} ref=A1,A2
CORREL sig=${eeg} ch-epoch ch-spatial-threshold=0.8 ch-high=0.98 '
Spatial weighting
The argument ch-spatial-weight only impacts the SUMM_MEAN
output, which is the mean of the absolute value of correlations for a
channel, with all other channels. (Note, this used not to take the
absolute value, but I've changed this behavior just now, along w/ the
other changes to CORREL).
As above, SUMM_MEAN may be based on epoch-level stats (after taking
the mean/median of those) or on whole-recording signals. In addition,
SUMM_MEAN is only based on below threshold channel pairs, if
ch-spatial-threshold has been set. Setting ch-spatial-weight will
weight the correlation, multiplying each absolute correlation by a
weight factor determine by the cosine similarity between the channel
pairs, before summing for that channel. The weight is set such that
more nearby channels have less weight -- i.e. if one is trying to pick
up on more distal channel pairs that still have high correlations in the signals.
Luna defines the weight as:
W = ( ( 1 - S ) / 2 )^X
i.e. we scale similarity S from ( -1 , +1 ) to ( 0 , 1 ) and then we
optionally take the X^th power, i.e. so that higher X values more
we put even less weight on nearby channel pairs. The default is 2.
e.g. for a linear effect, use:
ch-spatial-weight=1
CC
Calculates coupling and connectivity metrics
The CC command estimates cross-frequency coupling (currently
dPAC) and, for
inter-channel connectivity, the weighted phase lag index
(wPLI). It implements a
time-shifting randomization to generate the null distributions of
these metrics.
Note
Although CC currently only implements these two
metrics, in future releases other coupling and connectivity metrics
will be added within this framework (e.g. the coherence metrics
currently performed using COH).
For one of more signals, phase-amplitude coupling is estimated by the
dPAC method, if the pac option is specified. The phase and
amplitude of the signal is estimated using wavelets, with center
frequencies of fc and fc2 for phase and amplitude components
respectively, where fc is expected to be of lower frequency than
fc2. Multiple frequencies for either the phase (fc) or amplitude
(fc2) can be specified with a comma-delimited list, e.g. fc=1,2,4.
Alternatively, a grid of frequencies cab be specified by using
fc-range=1,20 instead of fc; here, it is also required to specify
num, e.g. num=20, which generates 20 fc values evenly spaced on
a log scale, between 1Hz and 20Hz. The linear option will force the grid
to be uniform on a linear, not log, scale instead. When considering phase-amplitude
coupling, only pairs where the phase frequency (fc) is lower than
the amplitude frequency (fc2) are retained for analysis.
As well as the wavelet center frequency, one can specify
the bandwidth of the wavelet, via the wavelet time-domain full width
at half maximum (FWHM), following this
convention.
See the CWT-DESIGN command for more
details, i.e. to better understand the implication of setting a
particular FWHM for the frequency domain properties of the wavelet.
In general, smaller FWHM values (in the time-domain) correspond to
broader FWHM in the frequency domain (i.e. a wider range of
frequencies above and below the center frequency will be captured).
Default FWHM values
If no FWHM values are specified, Luna will use a default value based on the frequency of the wavelet. For this, we seed on the default values selected in the Cox & Fell manuscript described in this vignette. In that manuscript, they used wavelet center frequencies of 0.5 to 30 Hz, evenly spaced on a log space, and paired these with 35 FWHM values evenly-spaced on a log scale from 5 to 0.25. For an arbitrary frequency, Luna will therefore use the following relation to generate a reasonable default value for the FWHM, which maintains equal frequency domain FWHM values on a log-scale:
exp(-0.7316762 * ln(F) + 1.1022791 )
Parameters
Core parameters
| Parameter | Example | Description |
|---|---|---|
sig |
sig=C3,C4 |
Optionally specify channels (default is to include all) |
fc |
fc=11,15 |
Specify wavelet center frequency/frequencies (phase) |
fwhm |
fwhm=1,1 |
Wavelet FWHM value(s) (phase) |
fc2 |
fc2=11,15 |
For PAC: as fc for amplitude |
fwhm2 |
fwhm2=1,1 |
For PAC: as fwhm for amplitude |
nreps |
nreps=1000 |
Number of randomized, surrogate time series generated |
pac |
pac |
Estimate within-channel phase-amplitude coupling metrics |
xch |
xch |
Estimate between-channel connectivity metrics |
Secondary parameters (primarily for specifying grids of frequency/FWHMs to test)
| Parameter | Example | Description |
|---|---|---|
fc-range |
fc-range=1,20 |
Specify range of center frequencies (requires num) (phase) |
fwhm-range |
fwhm-range=5,0.25 |
Specify range of FWHM values (requires num) (phase) |
num |
num=20 |
Number of intervals (evenly spaced on log scale) for fc-range or fwhm-range (phase) |
fc-range2 |
fc-range2=1,20 |
For PAC: as fc-range2 for amplitude |
fwhm-range2 |
fwhm-range2=5,0.25 |
For PAC: as fwhm-range2 for amplitude |
num2 |
num2=20 |
For PAC: as num for amplitude |
length |
length=10 |
Wavelet duration (in seconds) |
linear |
linear |
For ranges (e.g. fc-range) evenly space on a linear scale |
no-epoch-output |
no-epoch-output |
Suppress all epoch-level output |
That is, fc-range and num would be used instead of fc, etc.
Output
Whole-signal level metrics (strata: CH1 x CH2 x F1 x F2)
| Variable | Description |
|---|---|
CFC |
0/1 for whether this metric is a cross-frequency contrast (i.e. F1 != F2) |
XCH |
0/1 for whether this metric is a cross-channel contrast (i.e. CH1 != CH2 ) |
dPAC |
Debiased phase-amplitude coupling metric |
dPAC_Z |
Empirical Z-score for dPAC based on time-shuffling |
wPLI |
Weighted phase-lag index connectivity metric |
wPLI_Z |
Empirical Z-score for wPLI based on time-shuffling |
Epoch-level metrics (strata: E x CH1 x CH2 x F1 x F2)
| Variable | Description |
|---|---|
dPAC |
Debiased phase-amplitude coupling metric |
dPAC_Z |
Empirical Z-score for dPAC based on time-shuffling |
wPLI |
Weighted phase-lag index connectivity metric |
wPLI_Z |
Empirical Z-score for wPLI based on time-shuffling |
Example
See this vignette for an application of both dPAC and wPLI metrics.
Here, using two EEG channels from the second individual in the tutorial dataset, we extract only N2 sleep at consider wPLI between the two channels at a grid of 50 frequencies, linearly spaced between 1 and 25 Hz:
luna s.lst 2 -o out.db
-s 'MASK ifnot=NREM2 & RE &
CC sig=EEG,EEG(sec) fc-range=1,25 num=50 nreps=200 linear xch'
Extracting the output as follows:
destrat out.db +CC -r F1 F2 CH1 CH2
We can plot the wPLI (left) and correspondong Z-score (the empirical value derived from randomization, right):
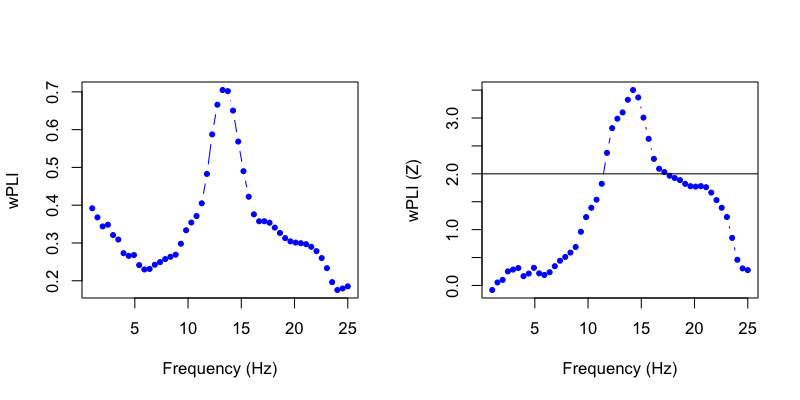
That is, we see significant connectivity between these two central channels in the spindle/sigma frequency band during N2 sleep.
Scaling the number of tests
Although this is designed to run reasonably efficiently, if you have lots of channels and/or lots of
frequencies and want to look at cross-frequency coupling between all pairs, then run time can get slow (...very slow...)
especially if you are using randomization to estimate Z scores (nreps) and/or have lots of individuals in the datasets.
Therefore, try to focus these analyses and start small...
PSI
Calculates the phase slope index across channels
Estimates the phase slop index between pairs of channels, and provides a single-channel summary of net PSI (i.e. net sender versus recipient).
The command requires that channels have similar sampling rates.
Parameters
To set the frequency or frequencies at which to calculate PSI either:
-
specify
fandw- one or more frequencies, for bandsf-w/2tof+w/2 -
or specify
f-lwr,f-upronly - for a band of lower to upper (or an equal range of comma-delimited values for each to give a range of bands, e.g.f-lwr=1,4,8andf-upr=4,8,12implies three bands: 1-4 Hz, 4-8 Hz and 8-12 Hz) -
or specify
f-lwr,f-upr,wandr- for a range of bands, fromfequallingf-lwrup tof-upr, in steps ofrHz; for each frequency, the window ofwspanning the center frequency is constructed
| Parameter | Example | Description |
|---|---|---|
sig |
sig=C3,C4 |
An optional parameter, to specify which channels to calculate PSI |
epoch |
Report epoch-level (e.g. 30-second epoch) output | |
f-lwr |
f-lwr=3 |
Consider frequencies from f-lwr to f-upr (as a band, or in steps of r ) |
f |
f=10,12,15 |
One or more frequencies to test (Hz) |
w |
w=5 |
Window around each center frequency (Hz) in which to calculate PSI |
Secondary parameters are:
| Parameter | Example | Description |
|---|---|---|
eplen |
eplen=5 |
PSI sub-epoch length (default 4 seconds) |
seglen |
seglen=2.5 |
Segment length (with 50% overlap) within each sub-epoch |
cache-metrics |
cache-metrics=c1 |
Cache PSI, e.g. for use with PSC |
Cache options
| Parameter | Example | Description |
|---|---|---|
cache-metrics |
cache-metrics=c1 |
Cache net and pairwsie PSC (e.g. for PSC) |
Output
Analysis parameter output (strata: F )
| Variable | Description |
|---|---|
F1 |
Lower frequency |
F2 |
Higher frequency |
NF |
Number of frequency bins |
Channel-level output (strata: CH )
| Variable | Description |
|---|---|
PSI |
Net PSI (standardized) for this channel |
Channel pair output (strata: CH1 x CH2)
| Variable | Description |
|---|---|
PSI |
Standardized PSI for this channel pair |
PSI_RAW |
Raw PSI |
STD |
Standard deviation of PSI |
Channel-level output (option: epoch, strata: E x CH )
| Variable | Description |
|---|---|
PSI |
Net PSI (standardized) for this channel |
Channel pair output (olption: epoch, strata: E x CH1 x CH2)
| Variable | Description |
|---|---|
PSI |
Standardized PSI for this channel pair |
PSI_RAW |
Raw PSI |
STD |
Standard deviation of PSI |
Example
See the walk-through for an example application of PSI.
XCORR
Efficient cross-correlation analysis
This command computes pairwise cross-correlations between signals using the FFT. All channels must have similar sample rates. This can be performed either epoch-wise, or on the whole signal. It computes correlations for a window of a fixed number of seconds w, optionally with an offset c (i.e. to find the maximum correlation of alignments from c-w to c+w seconds.
Parameters
| Parameter | Description |
|---|---|
sig |
Restrict analysis to these channels (at least two required) |
epoch |
Epoch-wise analysis |
w |
Optionally, maximum shift/window size to consider (seconds) |
c |
Optionally, offset to add (see above) (seconds) |
verbose |
Give verbose output |
Output
Pairwise channel measures for whole recording (option: epoch; strata: CH1 x CH2):
| Variable | Description |
|---|---|
D_MN |
Mean delay over epochs (seconds) |
D_MD |
Median delay over epochs (seconds) |
S_MN |
Mean delay over epochs (samples) |
S_MD |
Median delay over epochs (samples) |
D |
Delay based on average of lagged XCs (seconds) |
S |
Delay based on average of lagged XCs (samples) |
Pairwise cross-correlations for a given sample delay D (option:
verbose; strata: D x CH1 x CH2):
| Variable | Description |
|---|---|
T |
Lag in seconds |
XC |
Cross-correlation |
MI
Calculates pairwise mutual information metrics across channels
Estimates mutual information, a measure of dependence between two signals, based on methods described in Analyzing Neural Time Series Data by MX Cohen. Total correlation and dual total correlation are two normalized variants of the mutual information statistic: MI / min[ H(X), H(Y) ] and MI/H(X,Y) respectively, where MI is mutual information, H(X) and H(Y) are the marginal entropies, and H(X,Y) is the joint entropy.
Signals are first coarse-grained: the number of bins is determined by one of three rules: Freedman-Diaconis (default), Scott or Sturges rule, as described in Cohen.
Parameters
| Parameter | Example | Description |
|---|---|---|
sig |
sig=C3,C4,F3,F4 |
Optionally specify channels (default is to include all) |
epoch |
epoch |
Calculate and report MI and other measures per epoch |
scott |
scott |
Use Scott's rule to determine bin number |
sturges |
sturges |
Use Sturges' rule to determine bin number |
permute |
permute=1000 |
Estimate empirical significance via permutation, e.g. with 1000 null replicates |
Alert
Permutation and epoch-level analyses with the MI command can be relatively slow .
Output
Output for the whole signal (strata: CH1 x CH2)
| Variable | Description |
|---|---|
MI |
Mutual information |
TOTCORR |
Total correlation |
DTOTCORR |
Dual total correlation |
JINF |
Joint entropy |
INFA |
Marginal entropy of first signal |
INFB |
Marginal entropy of second signal |
NBINS |
Number of bins |
Output per-epoch, with epoch (option: epoch, strata: CH1 x CH2)
| Variable | Description |
|---|---|
MI |
Mutual information |
TOTCORR |
Total correlation |
DTOTCORR |
Dual total correlation |
JINF |
Joint entropy |
INFA |
Marginal entropy of first signal |
INFB |
Marginal entropy of second signal |
Output for permutation test (option: permute)
| Variable | Description |
|---|---|
EMP |
Empirical p-value for mutual information statistic |
Z |
Z-statistic |
TSYNC
Cross-correlation and phase delay
This function is now redundant - use XCORR instead
Estimate the cross-correlation between two signals, within a window of W seconds, and report the estimated phase delay (in seconds) based on the maximal cross-correlation in that time window.
Options
| Parameter | Example | Description |
|---|---|---|
sig |
sig=C3,C4,F3,F4 |
Optionally specify channels (default is to include all) |
w |
w=0.5 |
Required time window (seconds) |
verbose |
Verbose output | |
epoch |
Epoch-level output |
Output
Channel-pair output (strata: CH1 x CH2)
| Variable | Description |
|---|---|
S |
Phase delay based on cross-correlation |
Channel-pair output (option: verbose, strata: D x CH1 x CH2)
| Variable | Description |
|---|---|
XCORR |
Estimated cross-correlation for this delay |
Epoch-level channel-pair output (option: epoch, strata: CH1 x CH2)
| Variable | Description |
|---|---|
S |
Phase delay based on cross-correlation |
GP
Applies Granger prediction
(this section is a placeholder: the GP command is currently not fully tested/supported in the current Luna release)
Parameters
| Parameter | Example | Description |
|---|---|---|
sig |
sig=C3,C4,F3,F4 |
Optionally specify channels (default is to include all) |
Output
Channel-pair output (strata: CH1 x CH2)
| Variable | Description |
|---|---|
Example
to be added